Nuclear DNA Content
- The nucleus mainly have chromosome and nucleoplasm.
- The molecule of DNA in a single human chromosome ranges in size from 50 x 106 nucleotide pairs in the smallest chromosome (stretched full-length this molecule would extend 1.7 cm) up to 250 x 106 nucleotide pairs in the largest (which would extend 8.5 cm).
- Stretched end-to-end, the DNA in a single human diploid cell would extend over 2 meters. In the intact chromosome, however, this molecule is packed into a much more compact structure.
- The packing reaches its extreme during mitosis when a typical chromosome is condensed into a structure about 5 µm long (a 10,000-fold reduction in length).
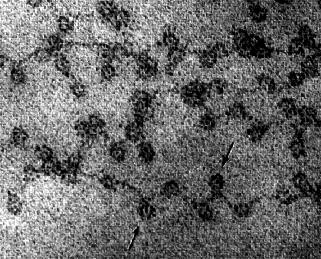
Figure 5.1: Electron micrograph showing chromatin from the nucleus of a chicken red blood cell (birds, unlike most mammals, retain the nucleus in their mature red blood cells). The arrows point to the nucleosomes. You can see why the arrangement of nucleosomes has been likened to “beads on a string”(courtesy of David E. Olins and Ada L. Olins).
Chromatin –
- The nucleus contains the chromosomes of the cell.
- Each chromosome consists of a single molecule of DNA complex with an equal mass of proteins. Collectively, the DNA of the nucleus with its associated proteins is called chromatin (Figure 5.1).
- Most of the protein consists of multiple copies of 5 kinds of histones.
- These are basic proteins, bristling with positively charged arginine and lysine residues. (Both Arg and Lys have a free amino group on their R group, which attracts protons (H+) giving them a positive charge.) Just the choice of amino acids you would make to bind tightly to the negatively-charged phosphate groups of DNA.
- Chromatin also contains small amounts of a wide variety of nonhistone proteins. Most of these are transcription factors (e.g., the steroid receptors) and their association with the DNA is more transient.
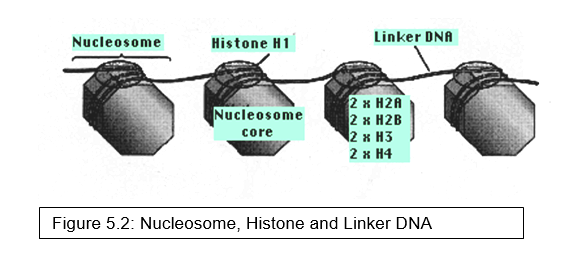
Nucleosomes – Two copies of each of four kinds of histones
- H2A
- H2B
- H3 and
- H4
form a core of protein, the nucleosome core (Figure 5.2).
- Around this is wrapped about 147 base pairs of DNA.
- From 20–60 bp of DNA link one nucleosome to the next. Each linker region is occupied by a single molecule of histone 1 (H1).
- The binding of histones to DNA does not depend on particular nucleotide sequences in the DNA but does depend critically on the amino acid sequence of the histone.
- Histones are some of the most conserved molecules during the course of evolution.
- Histone H4 in the calf differs from H4 in the pea plant at only 2 amino acids residues in the chain of 102.
- The formation of nucleosomes helps somewhat, but not nearly enough, to make the DNA sufficiently compact to fit in the nucleus.
- In order to fit 46 DNA molecules (in humans), totaling over 2 meters in length, into a nucleus that may be only 10 µm across requires more extensive folding and compaction.
- interactions between the exposed “tails” of the core histones cause nucleosomes to associate into a compact fiber 30 nm in diameter.
- these fibers are then folded into more complex structures whose precise configuration is uncertain and which probably changes with the level of activity of the genes in the region.
Histone Modifications –
- Although their amino acid sequence (primary structure) is unvarying, individual histone molecules do vary in structure as a result of chemical modifications that occur later to individual amino acids.
- These include adding:
- acetyl groups (CH3CO-) to lysines
- phosphate groups to serines and threonines
- methyl groups to lysines and arginines
- Although 75–80% of the histone molecule is incorporated in the core, the remainder – at the N-terminal – dangles out from the core as a “tail” (not shown in the Figure 5.2).
- The chemical modifications occur on these tails, especially of H3 and H4. Most of theses changes are reversible. For example, acetyl groups are
- added by enzymes called histone acetyltransferases (HATs)(not to be confused with the “HAT” medium used to make monoclonal antibodies!) and
- removed by histone deacetylases (HDACs).
- More often than not, acetylation of histone tails occurs in regions of chromatin that become active in gene transcription.
- This makes a kind of intuitive sense as adding acetyl groups neutralizes the positive charges on Lys thus reducing the strength of the association between the highly-negative DNA and the highly-positive histones.
- But there is surely more to the story.
- Acetylation of Lys-16 on H4 prevents the interaction of their “tails” needed to form the compact 30-nm structure of inactive chromatin.
- But this case involves interrupting protein-protein not protein-DNA interactions.
- Methylation, which also neutralizes the charge on lysines (and arginines), can either stimulate or inhibit gene transcription in that region.
- methylation of lysine-4 in H3 is associated with active genes while
- methylation of lysine-9 in H3 is associated with inactive genes. (These include those imprinted genes that have been permanently inactivated in somatic cells.) and
- Adding phosphates causes the chromosomes to become more – not less – compact as they get ready for mitosis and meiosis.
- In any case, it is now clear that histones are a dynamic component of chromatin and not simply inert DNA-packing material.
- Histone Variants – We have genes for 8 different varieties of histone 1(H1). Which variety is found at a particular linker depends on such factors as
- the type of cell,
- where it is in the cell cycle, and
- its stage of differentiation.
- In some cases, at least, a particular variant of H1 associates with certain transcription factors to bind to the enhancer of specific genes turning off expression of those genes.
Some other examples of histone variants:
- H3 is replaced by CENP-A (“centromere protein A”) at the nucleosomes near centromeres.
- Failure to substitute CENP-A for H3 in this regions blocks centromere structure and function.
- H2A may be replaced by the variant H2A.Z at the boundaries between euchromatin and heterochromatin.
- All the “standard” histones are replaced by variants as sperm develop.
- In general, the “standard” histones are incorporated into the nucleosomes as new DNA is synthesized during S phase of the cell cycle. Later, some are replaced by variant histones as conditions in the cell dictate.
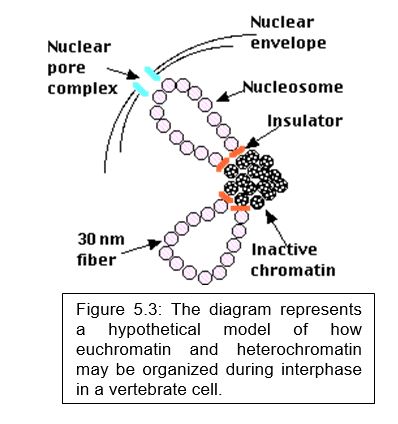
Euchromatin versus Heterochromatin –
- The density of the chromatin that makes up each chromosome (that is, how tightly it is packed) varies along the length of the chromosome.
- dense regions are called heterochromatin,
- less dense regions are called euchromatin (Figure 5.3).
Heterochromatin
- is found in parts of the chromosome where there are few or no genes, such as
- centromeres and
- telomeres
- is densely-packed;
- is greatly enriched with transposons and other “junk” DNA;
- is replicated late in S phase of the cell cycle;
- has reduced crossing over in meiosis.
- Those genes present in heterochromatin are generally inactive; that is, not transcribed and show
- increased methylation of the cytosines in CpG islands of the DNA;
- decreased acetylation of histones and
- increased methylation of lysine-9 in histone H3, which now provides a binding site for heterochromatin protein 1 (HP1), which blocks access by the transcription factors needed for gene transcription.
Euchromatin
- is found in parts of the chromosome that contain many genes;
- is loosely-packed in loops of 30-nm fibers.
- These are separated from adjacent heterochromatin by insulators.
- In yeast, the loops are often found near the nuclear pore complexes.
- This would seem to make sense making it easier for the gene transcripts to get to the cytosol.
- However, in animal cells, gene transcription appears to be repressed near the inner surface of the nuclear envelope.
- The genes in euchromatin are active and thus show decreased methylation of the cytosines in CpG islands of the DNA increased acetylation of histones and decreased methylation of lysine-9 in histone H3.
Nucleosomes and Transcription –
- Transcription factors cannot bind to their promoter if the promoter is blocked by a nucleosome.
- One of the first functions of the assembling transcription factors is to either expel the nucleosome from the site where transcription begins or at least to slide the nucleosomes along the DNA molecule.
- Either action exposes the gene’s promoter so that the transcription factors can then bind to it.
- The actual transcription of protein-coding genes is done by RNA polymerase II (RNAP II).
- In order for it to travel along the DNA to be transcribed, a complex of proteins removes the nucleosomes in front of it and then replaces them after RNAP II has transcribed that portion of DNA and moved on.
The Nucleolus –
- During the period between cell divisions, when the chromosomes are in their extended state, 1 or more of them (10 in human cells) have loops extending into a spherical mass called the nucleolus.
- Here are synthesized three (of the four) kinds of RNA molecules (28S, 18S, and 5.8S) used in the assembly of the large and small subunits of ribosome. 28S, 18S, and 5.8S ribosomal RNA is transcribed (by RNA polymerase I) from hundreds to thousands of tandemly-arranged rDNA genes distributed (in humans) on 10 different chromosomes.
- The rDNA-containing regions of these 10 chromosomes cluster together in the nucleolus. (In yeast, the 5S rRNA molecules – as well as transfer RNA molecules – are also synthesized (by RNA polymerase III) in the nucleolus.
- Once formed, rRNA molecules associate with the dozens of different ribosomal proteins used in the assembly of the large and small subunits of the ribosome.
- But all proteins are synthesized in the cytosol – and all the ribosomes are needed in the cytosol to do their work – so there must be a mechanism for the transport of these large structures in and out of the nucleus.
- This is one of the functions of the nuclear pore complexes.
Nucleoplasm –
- The term “nucleoplasm” is still used to describe the contents of the nucleus.
- However, the term disguises the structural complexity and order that seems to exist within the nucleus.
- For example, there is evidence that DNA replication and transcription occur at discrete sites within the nucleus.
- C-value is the mass of DNA (expressed, for example, in picograms per cell) in the haploid genome of a species.