![]()
Genomics
- Genomics is the study of whole genomes of organisms, and incorporates elements from genetics.
- Genomics uses a combination of recombinant DNA, DNA sequencing methods, and bioinformatics to sequence, assemble, and analyse the structure and function of genomes.
- It differs from ‘classical genetics’ in that it considers an organism’s full complement of hereditary material, rather than one gene or one gene product at a time.
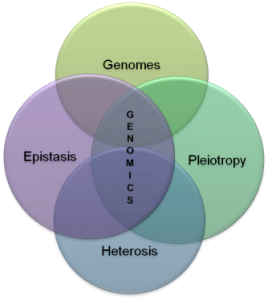
- Moreover, genomics focuses on interactions between loci and alleles within the genome and other interactions such as epistasis, pleiotropy and heterosis(Figure 1.1).
- Genomics harnesses the availability of complete DNA sequences for entire organisms and was made possible by both the pioneering work of Fred Sanger and the more recent next-generation sequencing technology.
- Fred Sanger’s group established techniques of sequencing, genome mapping, data storage, and bioinformatic analyses in the 1970s and 1980s.
- This work paved the way for the human genome project in the 1990s , an enormous feat of global collaboration that culminated in the publication of the complete human genome sequence in 2003.
- Today, next-generation sequence technologies have led to spectacular improvements in the speed, capacity and affordability of genome sequencing.
- Moreover, advances in bioinformatics have enabled hundreds of life-science databases and projects that provide support for scientific research.
- Information stored and organised in these databases can easily be searched, compared and analysed. We will explore some key genomics resources in the following sections of this course.
- Genomics is an interdisciplinary field of biology focusing on the structure, function, evolution, mapping, and editing of genomes.
- A genome is an organism’s complete set of DNA, including all of its genes.
- In contrast to genetics, which refers to the study of individual genes and their roles in inheritance, genomics aims at the collective characterization and quantification of all of an organism’s genes, their interrelations and influence on the organism.
- Genes may direct the production of proteins with the assistance of enzymes and messenger molecules.
- In turn, proteins make up body structures such as organs and tissues as well as control chemical reactions and carry signals between cells.
- Genomics also involves the sequencing and analysis of genomes through uses of high throughput DNA sequencing and bioinformatics to assemble and analyze the function and structure of entire genomes.
- Advances in genomics have triggered a revolution in discovery-based research and systems biology to facilitate understanding of even the most complex biological systems such as the brain.
- The field also includes studies of intragenomic (within the genome) phenomena such as epistasis (effect of one gene on another), pleiotropy (one gene affecting more than one trait), heterosis (hybrid vigour), and other interactions between loci and alleles within the genome.
Functional genomics
- Functional genomics is a field of molecular biology that attempts to make use of the vast wealth of data produced by genomic projects (such as genome sequencing projects) to describe gene (and protein) functions and interactions.
- Functional genomics focuses on the dynamic aspects such as gene transcription, translation, and protein–protein interactions, as opposed to the static aspects of the genomic information such as DNA sequence or structures.
- Functional genomics attempts to answer questions about the function of DNA at the levels of genes, RNA transcripts, and protein products.
- A key characteristic of functional genomics studies is their genome-wide approach to these questions, generally involving high-throughput methods rather than a more traditional “gene-by-gene” approach.
- A major branch of genomics is still concerned with sequencing the genomes of various organisms, but the knowledge of full genomes has created the possibility for the field of functional genomics, mainly concerned with patterns of gene expression during various conditions.
- The most important tools here are microarrays and bioinformatics.
Structural genomics
- Structural genomics seeks to describe the 3-dimensional structure of every protein encoded by a given genome.
- This genome-based approach allows for a high-throughput method of structure determination by a combination of experimental and modeling approaches.
- The principal difference between structural genomics and traditional structural prediction is that structural genomics attempts to determine the structure of every protein encoded by the genome, rather than focusing on one particular protein.
- With full-genome sequences available, structure prediction can be done more quickly through a combination of experimental and modeling approaches, especially because the availability of large numbers of sequenced genomes and previously solved protein structures allow scientists to model protein structure on the structures of previously solved homologs.
- Structural genomics involves taking a large number of approaches to structure determination, including experimental methods using genomic sequences or modeling-based approaches based on sequence or structural homology to a protein of known structure or based on chemical and physical principles for a protein with no homology to any known structure.
- As opposed to traditional structural biology, the determination of a protein structure through a structural genomics effort often (but not always) comes before anything is known regarding the protein function.
- This raises new challenges in structural bioinformatics, i.e. determining protein function from its 3D structure
Epigenomics
- Epigenomics is the study of the complete set of epigenetic modifications on the genetic material of a cell, known as the epigenome.
- Epigenetic modifications are reversible modifications on a cell’s DNA or histones that affect gene expression without altering the DNA sequence (Russell 2010 p. 475).
- Two of the most characterized epigenetic modifications are DNA methylation and histone modification.
- Epigenetic modifications play an important role in gene expression and regulation, and are involved in numerous cellular processes such as in differentiation/development and tumorigenesis.
- The study of epigenetics on a global level has been made possible only recently through the adaptation of genomic high-throughput assays.
Environmental Shotgun Sequencing (ESS) is a key technique in metagenomics.
(A) Sampling from habitat;
(B) filtering particles, typically by size;
(C) Lysis and DNA extraction;
(D) cloning and library construction;
(E) sequencing the clones;
(F) sequence assembly into contigs and scaffolds.
- Metagenomics is the study of metagenomes, genetic material recovered directly from environmental samples.
- The broad field may also be referred to as environmental genomics, ecogenomics or community genomics.
- While traditional microbiology and microbial genome sequencing rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample.
- Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods.
- Recent studies use “shotgun” Sanger sequencing or massively parallel pyrosequencing to get largely unbiased samples of all genes from all the members of the sampled communities.
- Because of its power to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world.
Model systems
Viruses and bacteriophages
- Bacteriophages have played and continue to play a key role in bacterial genetics and molecular biology.
- Historically, they were used to define gene structure and gene regulation.
- Also the first genome to be sequenced was a bacteriophage.
- However, bacteriophage research did not lead the genomics revolution, which is clearly dominated by bacterial genomics.
- Only very recently has the study of bacteriophage genomes become prominent, thereby enabling researchers to understand the mechanisms underlying phage evolution. Bacteriophage genome sequences can be obtained through direct sequencing of isolated bacteriophages, but can also be derived as part of microbial genomes.
- Analysis of bacterial genomes has shown that a substantial amount of microbial DNA consists of prophage sequences and prophage-like elements.
- A detailed database mining of these sequences offers insights into the role of prophages in shaping the bacterial genome:
- Overall, this method verified many known bacteriophage groups, making this a useful tool for predicting the relationships of prophages from bacterial genomes.
Cyanobacteria
- At present there are 24 cyanobacteria for which a total genome sequence is available. 15 of these cyanobacteria come from the marine environment.
- These are six Prochlorococcus strains, seven marine Synechococcus strains,
- Trichodesmium erythraeum IMS101 and Crocosphaera watsonii WH8501.
- Several studies have demonstrated how these sequences could be used very successfully to infer important ecological and physiological characteristics of marine cyanobacteria.
- However, there are many more genome projects currently in progress, amongst those there are further Prochlorococcus and marine Synechococcus isolates, Acaryochloris and Prochloron, the N2-fixing filamentous cyanobacteria Nodularia spumigena,
- Lyngbya aestuarii and Lyngbya majuscula, as well as bacteriophages infecting marine cyanobaceria.
- Thus, the growing body of genome information can also be tapped in a more general way to address global problems by applying a comparative approach.
- Some new and exciting examples of progress in this field are the identification of genes for regulatory RNAs, insights into the evolutionary origin of photosynthesis, or estimation of the contribution of horizontal gene transfer to the genomes that have been analyzed.
Applications of genomics
- Genomics has provided applications in many fields, including medicine, biotechnology, anthropology and other social sciences.
Genomic medicine
- Next-generation genomic technologies allow clinicians and biomedical researchers to drastically increase the amount of genomic data collected on large study populations.
- When combined with new informatics approaches that integrate many kinds of data with genomic data in disease research, this allows researchers to better understand the genetic bases of drug response and disease.
- For example, the All of Us research program aims to collect genome sequence data from 1 million participants to become a critical component of the precision medicine research platform.
Synthetic biology and bioengineering
- The growth of genomic knowledge has enabled increasingly sophisticated applications of synthetic biology.In 2010 researchers at the J. Craig Venter Institute announced the creation of a partially synthetic species of bacterium, Mycoplasma laboratorium, derived from the genome of Mycoplasma genitalium.
Conservation genomics
- Conservationists can use the information gathered by genomic sequencing in order to better evaluate genetic factors key to species conservation, such as the genetic diversity of a population or whether an individual is heterozygous for a recessive inherited genetic disorder.
- By using genomic data to evaluate the effects of evolutionary processes and to detect patterns in variation throughout a given population, conservationists can formulate plans to aid a given species without as many variables left unknown as those unaddressed by standard genetic approaches.