![]()
DNA Damage and Repair
Introduction to DNA Damage and Repair:
- DNA is a stable and versatile molecule.
- But sometimes the damage is caused to it,
- it is able to maintain stability by the information contained in it.
- DNA has many elaborate mechanisms to repair any damage or distortion.
- The most frequent sources of damage to DNA are the inaccuracy in DNA replication and chemical changes in DNA.
- Malfunction of the process of replication can lead to incorporation of wrong bases, which are mismatched with the complementary strand.
- The damage causing chemicals break the backbone of the strand and chemically alter the bases.
- Alkylation, oxidation and methylation cause damage to bases. X-rays and gamma radiations cause single or double stranded breaks in DNA.
- A change in the sequence of bases if replicated and passed on to the next generation becomes permanent and leads to mutation.
- At the same time mutations are also necessary which provide raw material for evolution.
- Without evolution, the new species, even human beings would not have arisen.
- Therefore a balance between mutation and repair is necessary.
Types of Damage:
Damage to DNA includes any deviation from the usual double helix structure.
1. Simple Mutations:
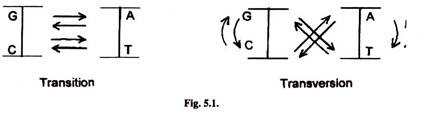
- Simplest mutations are switching of one base for another base.
- In transition one pyrimidine is substituted by another pyrimidine and purine with another purine.
- Trans-version involves substitution of a pyrimidine by a purine and purine by a pyrimidine such as T by G or A and A by C or T.
- Other simple mutations are detection, insertion of a single nucleotide or a small number of nucleotides.
- Mutations which change a single nucleotide are called point mutations.
2. Deamination:
- The common alteration of form or damage includes deamination of cytosine (C) to form uracil (u) which base pairs with adenine (A) in next replication instead of guanine (G) with which the original cytosine would have paired.
- As uracil is not present in DNA, adenine base pairs with thymine (T).
- Therefore the C-G pair is replaced by T-A in the next replication cycle. Similarly, hypoxanthine results from adenine deamination.
3. Missing Bases:
- Cleavage of the N-glycosidic bond between purine and sugar causes loss of purine base from DNA.
- This is called depurination.
- This apurinic site becomes a non-coding lesion.
4. Chemical Modification of Bases:
- Chemical modification of any of the four bases of DNA leads to modified bases.
- Methyl groups are added to various bases. Guanine forms 7- methylguanine, 3-methylguanine. Adenine forms 3-methyladenine.
- Cytosine forms 5- Methylcytosine.
- Replacement of amino group by a keto group converts 5-methylcytosine to thymine.
5. Formation of Pyrimidine Dimers (Thymine Dimers):
- Formation of thymine dimers is very common in which a covalent bond (cyclobutyl ring) is formed between adjacent thymine bases.
- This leads to loss of base pairing with opposite stand.
- A bacteria may have thousands of dimers immediately after exposure to ultraviolet radiations.
6. Strand Breaks:
- Sometimes phosphodiester bonds break in one strand of DNA helix.
- This is caused by various chemicals like peroxides, radiations and by enzymes like DNases.
- This leads to breaks in DNA backbone.
- Single strand breaks are more common than double strand breaks.
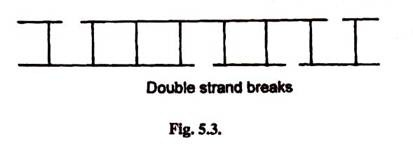
- Sometimes X-rays, electronic beams and other radiations may cause phosphodiester bonds breaks in both strands which may not be directly opposite to each other.
- This leads to double strand breaks.
- Some sites on DNA are more susceptible to damage. These are called hot-stops.
Repair Mechanisms:
- Most kinds of damage create impediments to replication or transcription.
- Altered bases cause mispairing and can cause permanent alteration to DNA sequence after replication.
- In order to maintain the integrity of information contained in it, the DNA has various repair mechanisms.
1. Direct Repair:
- The damage is reversed by a repair enzyme which is called photoreactivation.
- This mechanism involves a light dependent enzyme called DNA photolyase.
- The enzyme is present in almost all cells from bacteria to animals.
- It uses energy from the absorbed light to cleave the C-C bond of the cyclobutyl ring of the thymine dimers.
- In this way thymine dimers are monomerized.

2. Excision Repair:
It includes
- base excision repair (BER)
- nucleotide excision repair.(NER)
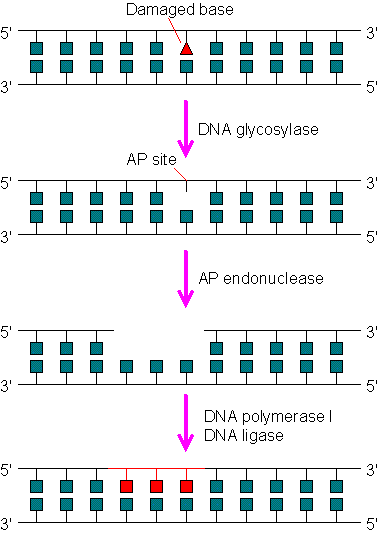
Base excision repair system (BER)
- It involves an enzyme called N-glycosylase which recognizes the abnormal base and hydrolyzes glycosidic bond between it and sugar.
- Another enzyme, an endonuclease cleaves the DNA backbone on the 5′-side of the abnormal base.
- Then the DNA polymerase by its exonuclease activity removes the abnormal base.
- DNA polymerase then replaces it with normal base and DNA ligase seals the region.

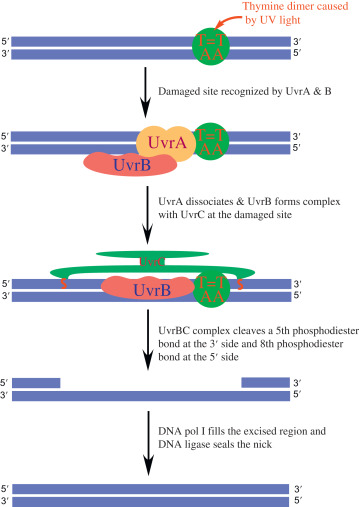
Nucleotide excision repair system (NER)
- It includes three steps, incision, excision and synthesis.
- Incision is done by endonuclease enzymes precisely on either side of the damaged patch of the strand.
- In this way the damaged portion of the strand is cleaved.
- Endonuclease enzymes involved are UvrA, UvrB which recognize the damaged stretch of the strand.
- UvrC makes two cuts (incisions) on either side.
- Exonuclease removes the damaged strand. Enzyme involved is UvrD.
- Later, DNA polymerase synthesizes the new strand by using complementary strands as a template.
- DNA ligase forms phosphodiester bonds which seal the ends on newly synthesized strands.
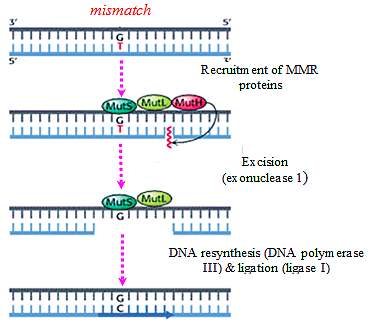
3. Mismatch Base Repair:
- Sometimes wrong bases are incorporated during the replication process, G-T or C-A pairs are formed.
- The wrong base is always incorporated in the daughter strand only.
- Therefore in order to distinguish the two strands for the purpose of repair, the adenine bases of the template strand are labeled or tagged by methyl groups.
- In this way the newly replicated DNA helix is hemimethylated.
- The excision of wrong bases occurs in the non-methylated or daughter strand.
- In this repair mechanism, three gene-products (proteins) are involved — Mut S, Mut L and Mut H.
- The first step of this repair process consists of binding of the Mut S protein to the mismatched base- pair.
- The second step involves the recognition of a specific sequence of the template which is -GATC- in E. coli in which A (adenine) is methylated in N-6 position.
- The proteins Mut L and Mut H which bind to the unmethylated -GATC- sequence of the new strand form a complex with Mut S which is bound to the mismatch pair.
- Thereby the mismatch pair is brought close to the -GATC- sequences.
- The Mut H protein then nicks the unmethylated DNA strand at the GATC site and the mismatch is removed by an exonuclease.
4. Recombination Repair or Retrieval System:
- In thymine dimer or other types of damage, DNA replication cannot proceed properly.
- A gap opposite to the thymine dimer is left in the newly synthesized daughter strand.
- The gap is repaired by a recombination mechanism or retrieval mechanism called sister strand exchange.
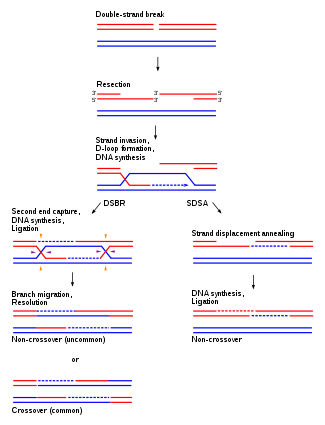
- During replication of DNA two identical copies are produced.
- Replicating DNA molecules has four strands A, B, C and D.
- Strands A and C have same DNA , sequence.
- Strands B and D also have same sequence as they are identical.
- A thymine dimer is present in strand A.
- The replication fork passes the dimmer as it cannot form hydrogen bonds with incoming adenine bases, thus creating a gap in the newly synthesized strand B.
- In recombination repair system a short identical segment of DNA is retrieved from strand D and is inserted into the gap of strand B.
- But this creates a gap in strand D which is easily filled up by DNA polymerase using normal strand C as a template.
- This event is dependent on the activity of a special protein Rec A.
- The Rec A protein plays its role in retrieving a portion of the complementary strand from other side of the replication fork to fill the gap.
- Rec A is a strand exchange protein.
- After filling both gaps, thymine is monomerised.
- So in this repair mechanism a portion of DNA strand is retrieved from the normal homologous DNA segment.
- This is also known as the daughter strand gap repair mechanism.
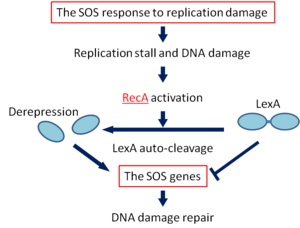
5. SOS Repair Mechanism:
- Sometimes the replication machinery is unable to repair the damaged portion and bypasses the damaged site.
- This is known as translesion synthesis also called bypass system and is an emergency repair system.
- This mechanism is catalyzed by a special class of DNA polymerases called Y-family of DNA polymerases which synthesize DNA directly across the damaged portion.
- The SOS response is set in action by the interaction of two proteins, — Rec A protein which is a product of the recA gene and Lex A protein, the product of lexA gene.
- The Rec A protein in addition to having a role in genetic recombination and recombinational repair also has a protease function.
- The Lex A protein acts as a repressor for a number of genes, known as SOS genes including the recA gene.
- Under normal conditions i.e. when the SOS response is not necessary, these genes remain repressed by the Lex A repressor.
- The initial event in the SOS response is the activation of RecA protease activity induced by DNA damage.
- The activation of Rec A protease activity occurs within a few minutes of DNA damage.
- The protease activity catalyzes cleavage of the Lex A repressor making it inactive.
- As a result, the SOS genes can now be expressed to produce the enzymes required for DNA repair.
- The SOS response, as the name suggests, is an emergency measure to repair mutational damage.
- It makes it possible for the cell to survive under conditions which would have been otherwise lethal.
- However, the possibility of generating new mutations increases in the repair of DNA molecules.
- This is because the SOS repair system allows DNA synthesis bypassing the damaged site.
- When the DNA polymerase III reaches a damaged site to which Rec A binds, the protein (Rec A) interacts with the epsilon subunit of the DNA polymerase molecule.
- This subunit is responsible for insertion of the correct base into the growing DNA strand. As a result, chain elongation continues bypassing the damaged site, but the chance of incorporation of a wrong base increases.
- SOS repair, therefore, enhances the chance of mutation due to mis-pairing of bases.
- This is known as error-prone bypass repair.
- A more recent model based on SOS repair of UV-irradiated DNA in bacteriophages has been proposed.
- UV-irradiation is known to produce dimers of not only thymine, but also of thymine and cytosine and cytosine.
- During replication when they are reached, the SOS repair system is halted temporarily and cytosine is deaminated to uracil.
- Uracil pairs with adenine, bringing about a transition mutation by changing C-G base pair to T-A.
- This is an error-free bypass repair though it still causes a mutation.
- It is called error-free because the template DNA strand is faithfully copied in the newly synthesized strand.
- The change from C to U occurs in the template strand itself.