![]()
STEPS OF DNA REPLICATION
DNA-Replication
Stages of Replication
The DNA synthesis or replication can be divided into three stages
(1) Initiation
(1) Elongation
(3) Termination
(i) Initiation :
- In E coli replication is origin a specific site which is called ori C.
- It is highly conserved sequence which is 245 bp long. .
- In this sequences there are three repeats of 13 bp AT rich sequence and four repeats of a 9 bp sequence.
- At least, nine different enzymes/ proteins participate in the initiation phase
- A single complex of four to five Dna A protein molecules binds to the four 9 bp repeats in the ori C
- DnaA protein forms a complex of 30-40 molecules, each bound to an ATP molecule and bacterial histone like proteins around which the OriC DNA becomes wrapped
- This process causes negatively supercoiling in DNA.
- This melting (unwoundıng) of three 13 bp AT-rich repeat sequences.
- After this Dna B with the help of Dna C protein loads on DNA.
- It is a hexamer ring structure with two rings.
- The two ring of Dna B, and DNA C complex loaded on to each DNA strand and unwinds the DNA bidirectionally by using energy of ATP hydrolysis to move into and melt double stranded DNA
- The single stranded bubble created in this way is coated with single strand binding protein (SSB) to protect it from breakage and prevent DNA renaturation.
- The enzyme DNA primase then attaches to the DNA and Synthesizes a short RNA primer to initiate synthesis of the leading stand of the first replication fork
- Bidirectional replication then follows
Unwinding :
- In replication DNA helicases travel along the template strands to open the double helix for copying the nucleotide on DNA template
- In addition to Dna B. a second DNA helicase may bind to the other strand to assist unwinding
- In a closed circular DNA molecule, removal of helical tums at the replication forks leads to the introduction of additional tums in the rest of the molecule in the form of positive supercoiling
- Although the natural negative supercoiling of circular DNA partially compensates for this, it is insufficient to allow continued progression of the replication forks
- This positive supercoiling must be relaxed continuously by the introduction of further negative supercoils by type Il topoisomerases (eg DNA gyrase)
(ii) Elongation :
- The elongation phase of replication includes two distinct but related events,
- leading strand synthesis and lagging strand synthesis
- Leading strand synthesis is more simple and begins with the synthesis of short (10 – 60 nucleotides) RNA primer by enzyme primase (Dna G protein)
- at the DNA- Replication Deoxyribonucleotides are added to this primer by DNA pol III.
i. Leading strand synthesis:
- Leading strand synthesis is a straightforward process which begins with the synthesis of RNA primer by primase at replication origin.
- DNA polymerase III then adds the nucleotides at 3’end.
- The leading strand synthesis then proceeds continuously keeping pace with unwinding of the replication fork until it encounters the termination sequences.
ii. Lagging strand synthesis:
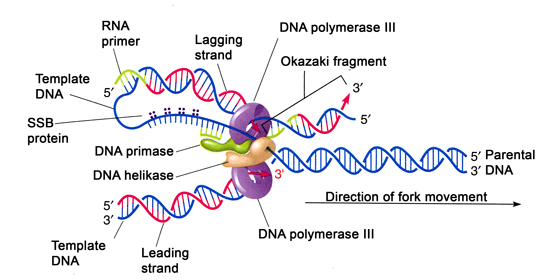
- The lagging strand synthesized in short fragments called Okazaki fragments.
- At first RNA primer is synthesized by primase and as in leading strand DNA polymerase III binds to RNA primer and adds dNTPS.
- On this level the synthesis of each okazaki fragments seems straight forward but the reality is quite complex.
Mechanism of Lagging strand synthesis
- The complexity lies in the coordination of leading and lagging strand synthesis.
- Both the strands are synthesized by a single DNA polymerase III dimer which accomplishes the looping of template DNA of lagging strand synthesizing Okazaki fragments.
- Helicase (dnaB) and primase (dnaG) constitute a functional unit within a replication complex called primosome.
- DNA pol III uses one set of core sub units (Core polymerase) to synthesize leading strand and another set of core subunit to synthesize lagging strand.
- In elongation steps, helicase in front of primase and pol III, unwind the DNA at the replication fork and travel along lagging strand templates along 5’-3’ direction.
- DnaG primase occasionally associated with dnaB helicase synthesizes short RNA primer. A new B-sliding clamp is then positioned at the primer by the B-clamp loading complex of DNA pol III.
- When the Okazaki fragments synthesis is completed, the replication halted and the core sub unit dissociates from their sliding clamps and associates with new clamp.
- This initiates the synthesis of new Okazaki fragments.
- Both leading and lagging strand are synthesized co-ordinately and simultaneously by a complex protein moving in 5’-3’ direction.
- In this way both leading and lagging strand can be replicated at same time by a complex protein that move in same direction.
- Every so often the lagging strands must dissociates from the replicosome and reposition itself so that replication can continue.
- Lagging strand synthesis is not completes until the RNA primer has been removed and the gap between adjacent Okazaki fragments are sealed.
- The RNA primer are removed by exonuclease activity (5’-3’) of DNA pol-I and replaced by DNA. The gap is then sealed by DNA ligase using NAD as cofactor.
(iii) Termination :
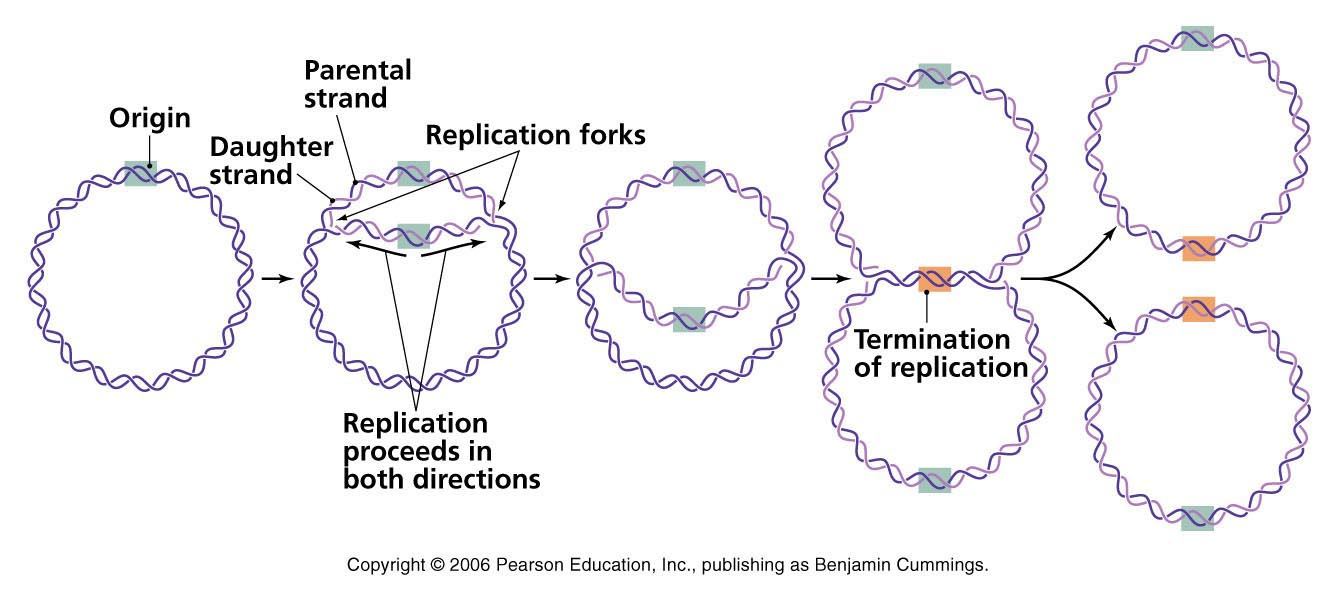
- Eventually the two replication forks of circular E. coli chromosomes meet at termination recognizing sequences (ter).
- The Ter sequence of 23 bp is arranged on the chromosome to create a trap that the replication fork can enter but cannot leave. Ter sequences function as binding sites for TUS protein.
- The Ter-TUS complex can arrest the replication fork from only one direction. Ter-TUS complex encounters first with either of the replication fork and halt it.
- The other opposing replication fork halted when it collided with the first one.
- This seems the Ter-TUS sequence is not essential for termination but it may prevent over replication by one fork if another is delayed or halted by a damage or some obstacle.
- When either fork encountered the Ter-TUS complex, replication halted.
- Final few hundred bases of DNA between these large protein complexes are replicated by not yet known mechanism forming two interlinked (catenated) chromosomes.
- In E. coli DNA topoisomerase IV (type II) cut the two strands of one circular DNA and segrate each of the circular DNA and finally join the strand.
- The DNA finally transferred to two daughter cells.



