![]()
REPLICATION
Semi-conservative replication of Chromosomes in eukaryotes:
- Autoradiography experiment in Vicia faba, by J.H. Taylor and his co-workers for the study of duplicating chromosomes in the root tip cells were first published in 1957.
- They reported that DNA in all the organisms has the inherent capacity of self-replication.
- The mechanism of DNA replication is so precise that all the cells derived from a zygote contain exactly similar DNA both in terms of quality and quantity.
- The replication takes place in interphase after every cell division.
Theoretically, there may be following three possible modes of DNA replication:
- Dispersive Method
- Conservative Method
- Semiconservative Method
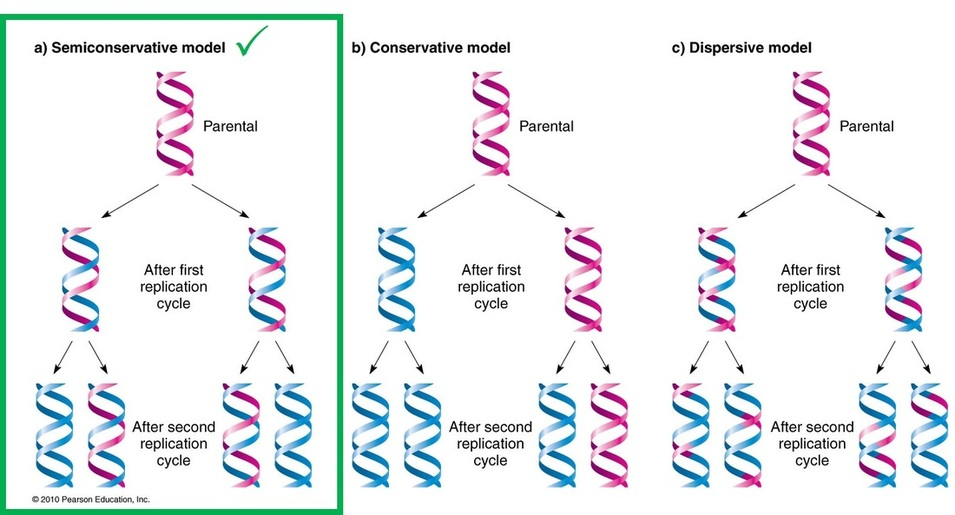
Semiconservative mode is the most accepted of all.
DNA Replication is Semi-Conservative:
- Watson and Crick model suggested that DNA replication is semi-conservative.
- It implies that half of the DNA is conserved.
- Only one new strand is synthesized, the other strand is the original DNA strand (template) that is retained.
- Each parental DNA strand serves as a template for one new complementary strand.
- The new strand is hydrogen bonded to its parental template strand and forms a double helix.
- Each of these strands of the double helix contains one original parental strand and one newly formed strand.
Meselson and Stahl Experiment:
- Mathew Meselson and Franklin Stahl proved experimentally that parental strands of a helix are distributed equally between the two daughter molecules.
- They made use of the heavy isotope 15N as a tag to differentially label the parental strands. E. coli was grown in a medium containing 15N labeled NH4Cl.
- In this way both strands of DNA molecules were labeled with radioactive heavy isotope 15N in their purines and pyrimidines.
- Therefore both strands were heavy or H DNA.
- The bacteria were then transferred into a medium containing the common non-radioactive nitrogen 14N, which is a light medium.
- It was found that after one cell division daughter molecules had one 15N strand the other 14N strand.
- So this is a hybrid molecule, a heavy light of HL.
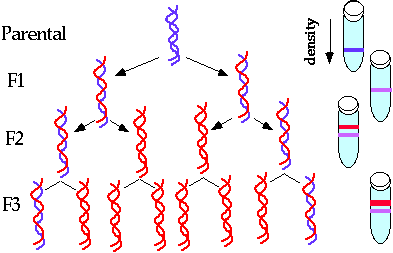
Fig. Semiconservative replication.
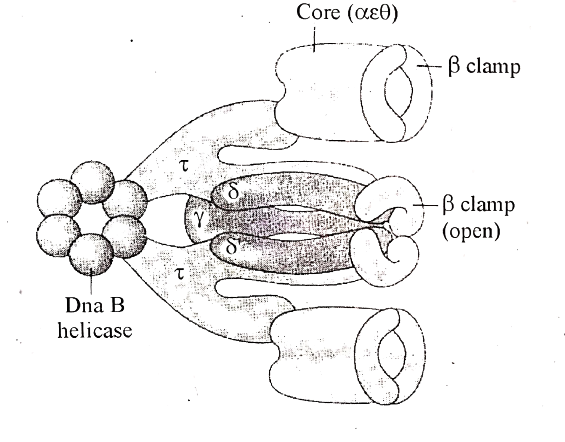
Enzymes Involved In DNA Replication:
Both the prokaryotic and eukaryotic cells contain three types of nuclear enzymes that are essential for DNA replication. These enzymes are nucleases, polymerases and ligases.
(i) Nucleases:
- The polynucleotide is held together by phosphodiester bonds.
- The nucleases hydrolyse the polynucleotide chain into the nucleotides.
- It attacks either at 3′ OH end or 5′ phosphate end of the chain.
- The nucleases are of two types.
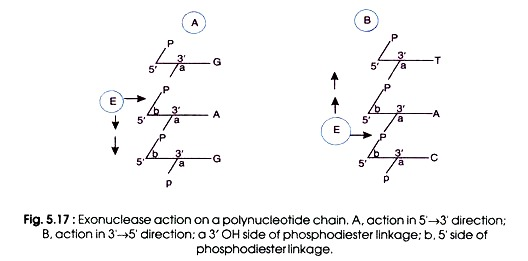
(a) Exonucleases:
- The nuclease that attacks the outer free end of the polynucleotide chain is called exonuclease.
- It breaks phosphodiester bonds either in direction 5’→ 3′ or in 3’→ 5′ direction.
- The enzyme moves in either cases stepwise along the chain and removes nucleotides one by one.
- Thus, the whole chain is digested.
(b) Endonucleases:
- The endonucleases attack within the inner portion of one or the double strands.
- A nick is made on a double stranded DNA molecule.
- However, if the polypeptide chain is single stranded (e.g. in DNA viruses), the attack of endonuclease will render the chain into two pieces.
- On double stranded DNA the nick contains two free ends that in turn act as template for DNA replication.
- Apart from this, the nicked double helix is distorted due to rotation of free molecules around its intact strand.
(ii) DNA Polymerases:
- DNA polymerases carry out the process of polymerization of nucleotides and formation of polynucleotide chains.
- This enzyme is called replicase when it replicates the DNA molecules and is inherited by daughter cells.
- In prokaryotes, three types of DNA polymerases e.g.
- polymerase I (Poly-I),
- polymerase II(Pol II), and
- DNA polymerase III (Pol III) are found,
- whereas in Eukaryotic cells contain five DNA polymerases: α, β, γ, δ, and ε. Polymerase γ is located in mitochondria and is responsible for replication of mitochondrial DNA.
- The other four enzymes are located in the nucleus and are therefore candidates for involvement in nuclear DNA replication.
- Polymerase β is most similar to E. coli DNA Pol I because its main function is associated with DNA repair, rather than replication. DNA polymerase β is mainly used in base-excision repair and nucleotide-excision repair.
- DNA polymerase 𝝳 is the main enzyme for replication in eukaryotes. It also has 3’→5′ exonuclease activity for proofreading. DNA polymerase 𝜶’is main function is to synthesize primers.
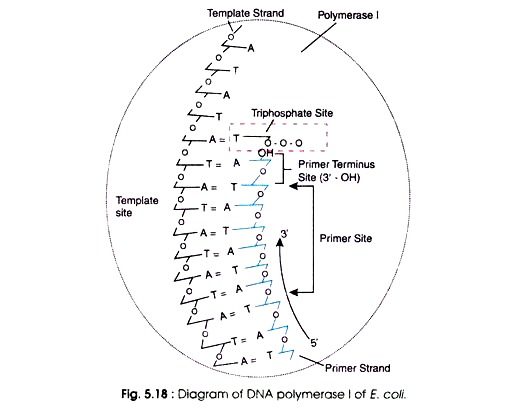
(a) Polymerase I (Pol I):
- It is discovered by The Kornberg

- It is also known as Kornberg polymerase .
- It is a single peptide chain with a molecular weight of 109,000 D.
- One atom of zinc (Zn) per chain is present, therefore, it is metalloenzyme.
- In E. coli, approximately 400 molecules of Pol I are present.
- It is roughly spherical of about 65 A° diameters.
Pol I possesses several attachment sites
such as:
(i) A template site for attachment to the DNA template,
(ii) A primer site of about 100 nucleotides contemporary to a segment of RNA on which the growth of newly synthesized DNA occur,
(iii) A primer terminus site containing a terminal 3’OH group at the tip, and
(iv) A triphosphate site for matching the incoming nucleoside triphosphates according to complementary nucleotide of DNA template.
Function:
In E.coli the following three types of functions of Pol I have been found.
Polymerization:
- it cannot synthesize a long chain.
- It synthesizes only a small segment of DNA in 5′ → 3′ direction
- it takes part in repair synthesis.
- In E.coli Pol I polymerize the nucleotides at the rate of 1,000 nucleotides per minute at 37°C.
Exonuclease activity:
3’ → 5′ exonuclease activity:
- Pol I catalyses the breaking of one or two DNA strands in 3’ → 5′ direction into the nucleotide components
- Therefore, it is called 3′ → 5′ exonuclease activity.
- Pol I correct the errors made during the polymerization, and edits the mismatching nucleotides at the primer terminus before the start of strand synthesis.
- Therefore, the function of Pol I is termed as repair synthesis.
5′ → 3′ exonuclease activity:
- Pol I also breaks the polynucleotide chain in 5′ → 3′ direction with the removal of nucleotide residues.
- Upon exposure of DNA to the ultraviolet light two adjacent pyrimidines such as thymines are covalently linked forming pyrimidine dimers.
- These dimers block the replication of DNA.
- Therefore, removal of pyrimidine dimers e.g. thymine dimers (T=T) is necessary.
- Through 5′ → 3′ exonuclease activity, Pol I removes pyrimidine dimers.
- Secondly, DNA synthesis occurs on RNA primer in the form Okazaki fragments.
- Through 5′ → 3′ exonuclease activity Pol I remove RNA primer and seal the gap with deoxyribonucleotides.
(b) Polymerase II (Pol II):
- Pol II is a single polypeptide chain (MW 90,000) that shows polymerization in 5’ → 3’ direction of a complementary chain.
- It also shows exonuclease activity in 3’ → 5’ direction but not in 5’ → 3’ direction.
- The polymerization activity of Pol II is much less than Pol I in E.coli cells.
- About 50 nucleotides per minute are synthesized.
- E.coli cells contain about 40 Pol II molecules.
- The 3′ → 5′ exonuclease activity of Pol II shows that it also plays a role in repair synthesis or DNA damaged by U.V. light just like Pol I.
- In the absence of Pol I, it can elongate the Okazaki fragments.
- Therefore, Pol II is an alternative to Pol I.
(c) Polymerase III (Pol III):
- DNA polymerase III is several times more active than Pol I and Pol II enzymes.
- It is the dimer of two polypeptide chains with molecular weight 1,40,000 and 40,000 D respectively.
- Pol III is the main polymerization enzyme that can polymerize about 15,000 nucleotides per minutes in E. coll.
- Synthesis of a long template also occurs when an auxiliary protein DNA (co-polymerase II) is linked with Pol III and produced Pol III-co Pol II complex.
- In addition Pol III also shows 3’→ 5′ exonuclease activity like Pol II.
- The 5’→ 3′ exonuclease activity is absent.

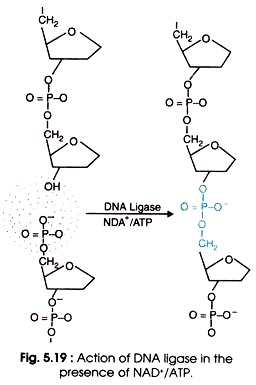
(iii) DNA Ligases:
- The DNA ligases seal single strand nicks in DNA which has 5’→ 3′ termini.
- It catalyses the formation of phosphodiester bonds between 3′-OH and 5′-PO4 group of a nick, and turns into an intact DNA.
- There are two types of DNA ligases: E. coli DNA ligase and T4 DNA ligase.
- The E. coli DNA ligase requires nicotinamide adenine dinucleotide (NAD+) as cofactor,
- whereas T4 DNA ligase uses ATP as cofactor for joining reaction of the nick (Fig 5.19).
DNA Helicase enzyme
- This is the enzyme that is involved in unwinding the double-helical structure of DNA allowing DNA replication to commence.
- It uses energy that is released during ATP hydrolysis, to break the hydrogen bond between the DNA bases and separate the strands.
- This forms two replication forks on each separated strand opening up in opposite directions.
- At each replication fork, the parental DNA strand must unwind exposing new sections of single-stranded templates.
- The helicase enzyme accurately unwinds the strands while maintaining the topography on the DNA molecule.
SSB proteins:
- Bind to the single strands of unwound DNA to prevent reformation of the DNA helix during replication.
DNA primase enzyme
- This is a type of RNA polymerase enzyme that is used to synthesize or generate RNA primers, which are short RNA molecules that act as templates for the initiation of DNA replication.
Topoisomerase
- This is the enzyme that solves the problem of the topological stress caused during unwinding.
- They cut one or both strands of the DNA allowing the strand to move around each other to release tension before it rejoins the ends.
- And therefore, the enzyme catalysts the reversible breakage it causes by joining the broken strands.
- Topoisomerase is also known as DNA gyrase in E. coli.



