Transposable Elements in Prokaryotes:
Although the presence of transposons was predicted in eukaryotes but first observation at molecular level was done in bacteria, There are four transposable genetic elements in prokaryotes:
(1) bacterial insertion sequences
(2) prokaryotic transposons
(3) insertion-sequence elements in plasmids and transposons and
(4) phage Mu.
(1) Bacterial insertion sequence:
Insertion sequence or insertion-sequence (IS) element:
- Insertion sequences, or insertion-sequence (IS) elements, are now known to be segments of bacterial DNA that can move from one position on one chromosome to a different position on the same chromosome or on a different chromosome.
- An IS element contains only the genes necessary to mobilize the element and insert the element into the chromosome at a new location.
- What elements are common components of bacterial chromosomes and plasmids?
- When IS elements appear in the middle of a gene, they disrupt the coding sequence and inactivate the expression of that gene.
- Due to their size and in some cases the presence of transcription and translation termination signals, IS elements can also block the expression of other genes in the same operon if they are downstream from the promoter of the gene operon.
- IS elements were first found in E. coli as a result of their effect on the expression of a set of three genes whose products are required to metabolize the sugar galactose as a carbon source.
- Careful investigation revealed that the mutant phenotype resulted from the insertion of a DNA segment of approximately 800 base pairs (bp) into a gene.
- This particular DNA segment is now called the insertion sequence (IS1).
Properties of IS Elements:
- Is1 is the genetic element capable of moving around the genome. It integrates into the chromosome at locations where it has no homologue, preventing it from recombination.
- This phenomenon is an example of transposition phenomenon.
- Several IS elements have been identified in E. coli, including IS1, IS2, and IS10, each present in 0 to 30 copies per genome, and each having a characteristic length and unique nucleotide sequence.
- IS1 is 768 bp long, and is present in 4 to 19 copies on E. coli chromosomes. IS2 is present in 0 to 12 copies on the E. coli chromosome and in one copy on the F plasmid, and IS10 is found in a class of plasmids called R plasmids that can replicate in E. coli

- In prokaryotes, IS elements are common cell components, that is, they are found in most cells. Overall, IS elements constitute approx. 0.3% of the cell’s genome.
- All IS elements sequenced end with complete or nearly complete inverted terminal duplication (IR) between 9 and 41 bp.
- This means that essentially the same sequence is found at each end of the IS but in the opposite orientation.
IS Transposition:
- When an IS element is translocated, a copy of the IS element inserts into a new chromosomal location, while the original IS element remains intact.
- That is, transposition requires exact replication of the original IS element using replication enzymes of the host cell.
- The actual transposition also requires an enzyme encoded by the IS element called a transposase.
- IR sequences are essential for the transposition process, that is, those sequences are recognized by transposase to initiate transposition.
- Are elements inserted into chromosomes at locations where they have no sequence homology?
- Genetic recombination between non-homologous sequences is called illegitimate recombination.
- The sites that contain IS elements are called target sites.
- The process of IS insertion into the chromosome is shown in Figure
- First, a tag is cut into the target site and then the IS element is inserted, which binds to the zigzag-stranded ends.
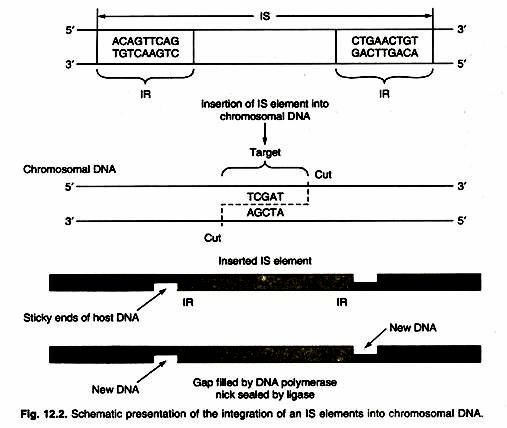
- The gap is filled by DNA polymerase and DNA ligase, creating an integrated IS element with two direct repeats of the target site sequence flanking the IS element. ‘Direct’ in this case means that the two sequences are repeated in the same orientation.
- Direct duplication is called target site duplication.
- The size of target site repeats varies with IS elements, but is small.
- Integration of some IS elements shows a preference for certain areas, while others are integrated only in particular sequences.
- All copies of a given IS element have the same sequence, including inverted terminal repeats.
- Mutations that affect the inverted terminal repeat sequence of IS elements affect transposition, indicating that inverted terminal repeat sequences are the major sequences recognized by transposase during the transposition event.
(2) Prokaryotic transposon:
- A transposon (Tn) is more complex than IS elements.
- A transposon is a mobile DNA segment that, like an IS element, contains genes to insert the DNA segment into the chromosome and to assemble the element at other locations on the chromosome.
- There are two types of prokaryotic transposons: mixed transposons and non- composite transposons
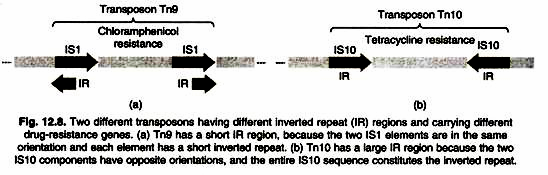
(1) Composite transposon:
- They are complex transposons consisting of a central region containing genes, for example, drug resistance genes, surrounded on both sides by IS elements (also called IS modules).
- Mixed transposons can be thousands of base pairs long.
- The IS elements are both of the same type and are called IS-L (for “left”) and IS-R (for “right”).
- Depending on the transposon, IS-L and IS-R may be in the same or inverted orientation relative to each other.
- Because the IS itself contains terminal inverted repeats, the overall transposon also contains terminal inverted repeats.
- Figure shows the structure of the compound transposon Tn10 to illustrate the general characteristics of such transposons.
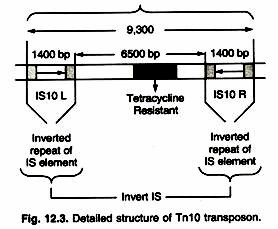
- The TN10 transposon is 9,300 bp long and consists of 6,500 bp of central, non-repetitive DNA containing the tetracycline resistance gene flanked by a 1,400-bp IS element at each end.
- These IS elements are designated IS10L and IS10R and are arranged in an inverted orientation.
- Cells containing TN10 are resistant to the tetracycline resistance gene contained in the central DNA sequence.
- Transfer of mixed transposons occurs due to the function of IS elements present in them.
- One or both IS elements supply transposase.
- Inverted repeats of IS elements at both ends of the transposon are recognized by the transposase to initiate transposition (as occurs with transposition of IS elements).
- Translocation of TN10 is rare, occurring once in 10 cell generations.
- This is because less than one transposase molecule is produced by Tn 10 per cell generation.
- Like IS elements, mixed transposons generate target site duplications after transposition.
(2) Non-mixed transposons:
- They like mixed transposons, they contain genes like drug resistance.
- Unlike mixed transposons, they do not end with IS elements.
- However, they contain repeated sequences at their ends that are essential for translocation.
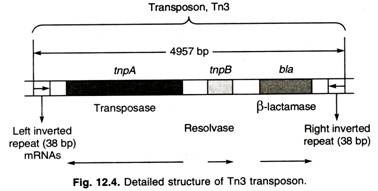
- Tn3 is a non-mixed transposon.
- Tn3 has 38 bp inverted terminal repeats and three genes in its central region.
- One of those genes, bla, encodes a β-lactamase that breaks down ampicillin and therefore makes Tn3-containing cells resistant to ampicillin.
- The other two genes, tnpA and tnpB, encode the enzymes transposase and resolvase that are required for transposition of Tn3 (Figure ).
- Transposase catalyzes the insertion of TNs into new sites, and resolvase is an enzyme involved in specialized recombination events associated with transposition.
- Resolvase is not found in all transposons.
- The genes for transposition are in the central region for non-composite transposons, whereas they are in the terminal IS elements for composite transposons.
- Non-mixed transposons also cause target site duplication when they move.
(3) Mechanism of transposition in prokaryotes:
- Several different mechanisms of transposition are employed by prokaryotic transposable elements. And, as we will see later, eukaryotic elements exhibit even additional mechanisms of transfer.
- In E. coli, we can identify replicative and conservative (non-replicative) modes of transposition.
- In the replication pathway, a new copy of the transposable element is generated in a transposition event.
- The result of transposition is that one copy appears at the new site and one copy remains at the old site.
- In the Orthodox path, there is no replication. Instead, the element is removed from the chromosome or plasmid and integrated into the new site (Figure ).
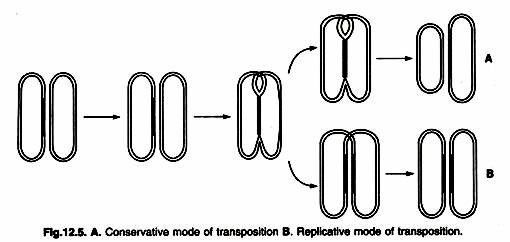
Replica Transfer:
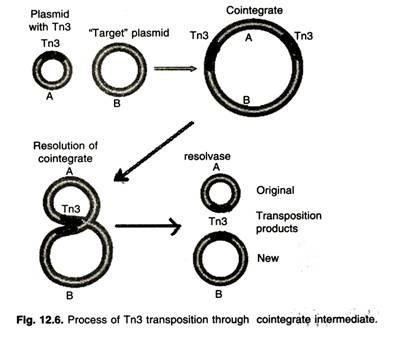
- The transfer of Tn3 occurs in two stages. First, the transposase mediates the fusion of the two molecules, forming a structure called a coinintegrate.
- During this process, the transposon is replicated, and one copy is inserted at each junction in the co-integrator.
- Two Tn3 are oriented in the same direction. In the second step of transposition, the tnpR-encoded resolvase mediates a site-specific recombination event between two Tn3 elements.
- This event occurs in Tn3 at a sequence called the res, the resolution site, and generates two molecules, each containing a copy of the transposon.
- The TNPR gene-product also has another function, namely, to suppress the synthesis of both transposase and resolvase proteins.
- This repression occurs because the race site is located between the tnpA and tnpr genes.
- By binding to this site, the tnpR protein interferes with the synthesis of both gene-products, reducing their supply.
- As a result, the Tn3 element remains stable (Figure ).
Conservative transfers:
- Some transposons, such as Tn10, are released from the chromosome and integrate into the target DNA. In these cases,
- DNA replication of the element does not occur, and the element is lost from the site of the original chromosome.
- This mechanism is called conservative (nonreplicative) transposition or simple insertion. TN10, for example, transfers by conservative transfers.
- Inserting a transposon into the reading frame of a gene will disrupt it, causing loss of function of that gene.
- Entry into the control region of a gene can lead to changes in gene expression levels.
- Deletion and insertion events also occur as a result of the activities of transposons and as a result of crossing-over between duplicate transposons in the genome.
(3) IS elements and transposons in plasmids:
- The transfer of genetic material between conjugal E. coli is the result of the action of reproduction factor F.
- The F factor, a circular double-stranded DNA molecule, is one of the examples of bacterial plasmids. F-like plasmids that are also capable of integrating into bacterial chromosomes are called episomes.
- F factor consists of 94,500 bp of DNA that codes for a variety of proteins.
The important elements are:
(i) Transfer gene (Tra) is essential for conjugative transfer of DNA.
(ii) Genes that encode proteins required for replication of the plasmid,
(iii) Four IS elements, two copies of IS3, one of IS2, and one of an insertion sequence element called gamma-delta.
- This is because there are copies of these four insertion sequences at different locations in the E. coli chromosome, which the F factor can integrate into the E. coli chromosome at different locations and in different orientations with the homologous sequence of the insertion elements.
- Another class of plasmids that has therapeutic importance is the R plasmid group, which was discovered in Japan in the 1950s during the treatment of dysentery.
- This disease is the result of infection with the pathogenic bacterium Shigella. Shigella was found to be resistant to most commonly used antibiotics.
- Subsequently, they discovered that the genes responsible for drug resistance were carried on R plasmids, which, like the F factor, could promote the transfer of genes between bacteria by conjugation.
- A segment of the R plasmid that is homologous to a segment in the F factor is the part required for marital transfer of the gene.
- That segment and the plasmid-specific genes for DNA replication are called the RTF (resistance transfer factor) region (Figure 12.7).
- The remainder of the R plasmid varies in type and may contain antibiotic-resistance genes or other types of genes of medical importance, such as resistance to heavy metal ions.
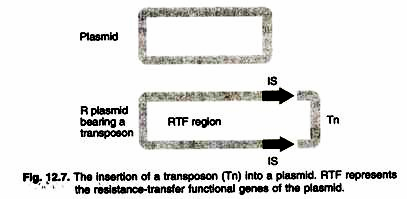
- Resistance genes in R plasmids are, in fact, transposons, i.e. each resistance gene is located between flanking, directly repeated segments such as one of the IS modules (Fig. ).
- Thus, each transposon with its resistance gene in the R plasmid can be inserted into a new location on another plasmid or on the bacterial chromosome, while at the same time leaving a copy of itself in the original position.
(4) Phase Mu:
- Phage Mu is a normal appearing phage. We are considering it here because, although it is a true virus, it has many characteristics in common with IS elements.
- The DNA double helix of this phage is 36,000 nucleotides long – much larger than the IS element.
- However, it appears that it is capable of inserting itself anywhere in the bacterial or plasmid genome in any direction.
- Once inserted, it causes mutations at the site of insertion—again like an IS element. (The phage was named for this ability: mu means “mutant”.)
- Generally, these mutations cannot be undone, but reversion can be produced by some type of genetic manipulation.
- When this reversion occurs, the phages that can be recovered show no deletions, proving that the excision is accurate and therefore the insertion of the phage causes no loss of phage material.
- Each mature phage particle has a flanking DNA fragment from its previous host at each end.
- However, this DNA is not inserted de novo into the next host. Its function is unclear. Phage Mu also has an IR sequence, but none of the repeated elements are at the terminus.

- Mu can also act like a genetic snap fastener, mobilizing any type of DNA and transferring it anywhere in the genome.
- For example, it may mobilize another phase (such as λ) or F factor.
- In such situations, the inserted DNA is surrounded by two Mu genomes.
.