Site Specific Recombination
Site-specific Recombination Enzymes Move Special DNA Sequences into and out of Genomes
- Site-specific genetic recombination, unlike general recombination, is guided by a recombination enzyme that recognizes specific nucleotide sequences present on one or both of the recombining DNA molecules.
- Base-pairing between the recombining DNA molecules need not be involved, and even when it is, the heteroduplex joint that is formed is only a few base pairs long.
- By separating and joining double-stranded DNA molecules at specific sites, this type of recombination enables various types of mobile DNA sequences to move about within and between chromosomes.
- Site-specific recombination was first discovered as the means by which a bacterial virus, bacteriophage lambda, moves its genome into and out of the E. coli chromosome. In its integrated state the virus is hidden in the bacterial chromosome and replicated as part of the host’s DNA.
- When the virus enters a cell, a virus-encoded enzyme called lambda integrase is synthesized.
- This enzyme catalyzes a recombination process that begins when several molecules of the integrase protein bind tightly to a specific DNA sequence on the circular bacteriophage chromosome.
- The resulting DNA-protein complex can now bind to a related but different specific DNA sequence on the bacterial chromosome, bringing the bacterial and bacteriophage chromosomes close together.
- The integrase then catalyzes the required DNA cutting and resealing reactions, using a short region of sequence homology to form a tiny heteroduplex joint at the point of union (Figure 3.18).

fig 3.18
- The integrase resembles a DNA topoisomerase in that it forms a reversible covalent linkage to DNA wherever it breaks a DNA chain.
- The same type of site-specific recombination mechanism can also be carried out in reverse by the lambda bacteriophage, enabling it to exit from its integration site in the E. coli chromosome in order to multiply rapidly within the bacterial cell.
- This excision reaction is catalyzed by a complex of the integrase enzyme with a second bacteriophage protein, which is produced by the virus only when its host cell is stressed. If the sites recognized by such a recombination enzyme are flipped, the DNA between them will be inverted rather than excised.
- Many other enzymes that catalyze site-specific recombination resemble lambda integrase in requiring a short region of identical DNA sequence on the two regions of DNA helix to be joined.
- Because of this requirement, each enzyme in this class is fastidious with respect to the DNA sequences that it recombines, and it can be expected to catalyze one particular DNA joining event that is useful to the virus, plasmid, transposable element, or cell that contains it.
- These enzymes can be exploited as tools in transgenic animals to study the influence of specific genes on cell behavior, .
- Site-specific recombination enzymes that break and rejoin two DNA double helices at specific sequences on each DNA molecule often do so in a reversible way: as for lambda bacteriophage, the same enzyme system that joins two DNA molecules can take them apart again, precisely restoring the sequences of the two original DNA molecules.
- This type of recombination is therefore called conservative site-specific recombination to distinguish it from the mechanistically distinct transpositional site-specific recombination that we discuss next.
Transpositional Recombination Can Insert a Mobile Genetic Element into Any DNA Sequence
- Many mobile DNA sequences, including many viruses and transposable elements, encode integrases that insert their DNA into a chromosome by a mechanism that is different from that used by bacteriophage lambda.
- Like the lambda integrase, each of these enzymes recognizes a specific DNA sequence in the particular mobile genetic element whose recombination it catalyzes.
- Unlike the lambda enzyme, however, these integrases do not require a specific DNA sequence in the “target” chromosome and they do not form a heteroduplex joint.
- Instead, they introduce cuts into both ends of the linear DNA sequence of the mobile genetic element and then catalyze a direct attack by these DNA ends on the target DNA molecule, breaking two closely spaced phosphodiester bonds in the target molecule.
- Because of the way that these breaks are made, two short single-stranded gaps are left in the recombinant DNA molecule, one at each end of the mobile element; these are filled in by DNA polymerase to complete the recombination process. As illustrated in Figure 3.20,
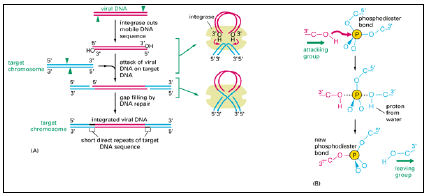
fig 3.20
- this mechanism creates a short duplication of the adjacent target DNA sequence; such flanking duplications are the hallmark of a transpositional site-specific recombination event.
- An integrase enzyme of this type was first purified in active form from bacteriophage Mu.
- Like the bacteriophage lambda integrase, it carries out all of its cutting and rejoining reactions without requiring an energy source (such as ATP). Very similar enzymes are present in organisms as diverse as bacteria, fruit flies, and humans – all of which contain mobile genetic elements, as we discuss next.



