Regulation of Gene Expression in Prokaryotes and Eukaryotes
- While the period from 1900 to the Second World War has been called the “golden age of genetics”, we may be in a new golden (or platinum) age.
- Recombinant DNA technology allows us to manipulate the very DNA of living organisms and to make conscious changes in that DNA.
- Prokaryote genetic systems are much easier to study and better understood than are eukaryote systems.
Gene Regulation in Prokaryotes
(1) In Bacteria –
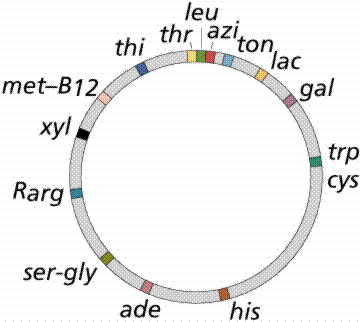
- The single chromosome of the common intestinal bacterium E. coli is circular and contains some 4.7 million base pairs. It is nearly 1 mm long, but only 2nm wide (Figure 2.32).
- The chromosome replicates in a bidirectional method, producing a figure resembling the Greek letter theta.
- The promoter is the part of the DNA to which the RNA polymerase binds before opening the segment of the DNA to be transcribed.
- A segment of the DNA that codes for a specific polypeptide is known as a structural gene.
- These often occur together on a bacterial chromosome.
- The location of the polypeptides, which may be enzymes involved in a biochemical pathway, for example, allows for quick, efficient transcription of the mRNAs.
- Often leader and trailer sequences, which are not translated, occur at the beginning and end of the region. E. coli can synthesize 1700 enzymes.
- Therefore, this small bacterium has the genes for 1700 different mRNAs.
The Operon Model –
- The operon model (Figure 2.33) of prokaryotic gene regulation was proposed by Fancois Jacob and Jacques Monod. Groups of genes coding for related proteins are arranged in units known as operons.
- An operon consists of an operator, promoter, regulator, and structural genes.
- The regulator gene codes for a repressor protein that binds to the operator, obstructing the promoter (thus, transcription) of the structural genes.
- The regulator does not have to be adjacent to other genes in the operon. If the repressor protein is removed, transcription may occur.
- Operons are either inducible or repressible according to the control mechanism.
- Seventy-five different operons controlling 250 structural genes have been identified for E. coli. Both repression and induction are examples of negative control since the repressor proteins turn off transcription.
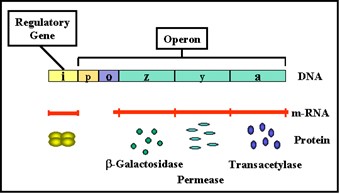
Figure 2.33 : The lactose operon
- Bacteria do not make all the proteins that they are capable of making all of the time.
- Rather, they can adapt to their environment and make only those gene products that are essential for them to survive in a particular environment.
- For example, bacteria do not synthesize the enzymes needed to make tryptophan when there is an abundant supply of tryptophan in the environment.
- However, when tryptophan is absent from the environment the enzymes are made. Similarly, just because a bacterium has a gene for resistance to an antibiotic does not mean that that gene will be expressed.
- The resistance gene may only be expressed when the antibiotic is present in the environment.
- Bacteria usually control gene expression by regulating the level of mRNA transcription.
- In bacteria, genes with related function are generally located adjacent to each other and they are regulated coordinately (i.e. when one is expressed, they all are expressed).
- Coordinate regulation of clustered genes is accomplished by regulating the production of a polycistronic mRNA (i.e. a large mRNA containing the information for several genes).
- Thus, bacteria are able to “sense” their environment and express the appropriate set of genes needed for that environment by regulating transcription of those genes.
(A). INDUCIBLE GENES – THE OPERON MODEL
1. Definition
- An inducible gene is a gene that is expressed in the presence of a substance (an inducer) in the environment.
- This substance can control the expression of one or more genes (structural genes) involved in the metabolism of that substance.
- For example, lactose induces the expression of the lac genes that are involved in lactose metabolism. An certain antibiotic may induce the expression of a gene that leads to resistance to that antibiotic.
- Induction is common in metabolic pathways that result in the catabolism of a substance and the inducer is normally the substrate for the pathway.
2. Lactose Operon
a. Structural genes –
- The lactose operon (Figure 2.33) contains three structural genes that code for enzymes involved in lactose metabolism.
- The lac z gene codes for β-galactosidase, an enzyme that breaks down lactose into glucose and galactose
- The lac y gene codes for a permease, which is involved in uptake of lactose
- The lac a gene codes for a galactose transacetylase.
- These genes are transcribed from a common promoter into a polycistronic mRNA, which is translated to yield the three enzymes.
b. Regulatory gene –
- The expression of the structural genes is not only influenced by the presence or absence of the inducer, it is also controlled by a specific regulatory gene.
- The regulatory gene may be next to or far from the genes that are being regulated.
- The regulatory gene codes for a specific protein product called a REPRESSOR.
c. Operator –
- The repressor acts by binding to a specific region of the DNA called the operator which is adjacent to the structural genes being regulated.
- The structural genes together with the operator region and the promoter is called an OPERON.
- However, the binding of the repressor to the operator is prevented by the inducer and the inducer can also remove repressor that has already bound to the operator.
- Thus, in the presence of the inducer the repressor is inactive and does not bind to the operator, resulting in transcription of the structural genes.
- In contrast, in the absence of inducer the repressor is active and binds to the operator, resulting in inhibition of transcription of the structural genes.
- This kind of control is referred to a NEGATIVE CONTROL since the function of the regulatory gene product (repressor) is to turn off transcription of the structural genes.
d. Inducer –
- Transcription of the lac genes is influenced by the presence or absence of an inducer (lactose or other β-galactosides) (Figure 2.34).
- e.g:- + inducer = expression and – inducer = no expression
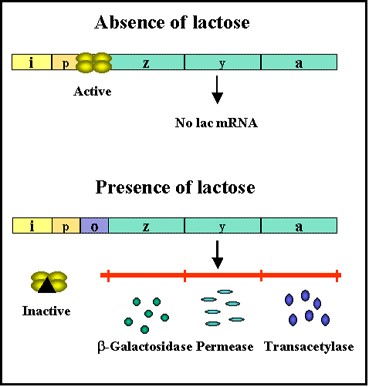
Figure 2.34 : Transcription of lac genes in the presence and absence of glucose
3. Catabolite repression (Glucose Effect)
- Many inducible operons are not only controlled by their respective inducers and regulatory genes, but they are also controlled by the level of glucose in the environment.
- The ability of glucose to control the expression of a number of different inducible operons is called CATABOLITE REPRESSION.
- Catabolite repression is generally seen in those operons which are involved in the degradation of compounds used as a source of energy.
- Since glucose is the preferred energy source in bacteria, the ability of glucose to regulate the expression of other operons ensures that bacteria will utilize glucose before any other carbon source as a source of energy.
Mechanism –
- There is an inverse relationship between glucose levels and cyclic AMP (cAMP) levels in bacteria.
- When glucose levels are high cAMP levels are low and when glucose levels are low cAMP levels are high.
- This relationship exists because the transport of glucose into the cell inhibits the enzyme adenyl cyclase which produces cAMP.
- In the bacterial cell cAMP binds to a cAMP binding protein called CAP or CRP.
- The cAMP-CAP complex, but not free CAP protein, binds to a site in the promoters of catabolite repression-sensitive operons.
- The binding of the complex results in a more efficient promoter and thus more initiations of transcriptions from that promoter as illustrated in Figures 2.35 and 2.36.
- Since the role of the CAP-cAMP complex is to turn on transcription this type of control is said to be POSITIVE CONTROL.
- The consequences of this type of control is that to achieve maximal expression of a catabolite repression sensitive operon glucose must be absent from the environment and the inducer of the operon must be present.
- If both are present, the operon will not be maximally expressed until glucose is metabolized. Obviously, no expression of the operon will occur unless the inducer is present.

Figure 2.35 : Effect of glucose on expression of proteins encoded by the lac operon
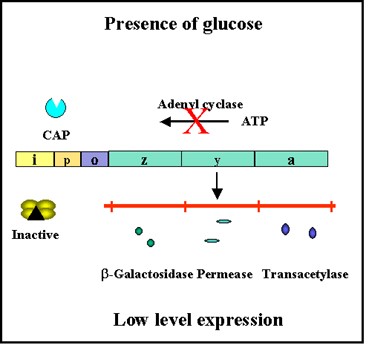
Figure 2.36 : Effect of glucose on expression of proteins encoded by the lac operon
(B). REPRESSIBLE GENES – THE OPERON MODEL
1. Definition
- Repressible genes are those in which the presence of a substance (a co-repressor) in the environment turns off the expression of those genes (structural genes) involved in the metabolism of that substance. e.g.,
- Tryptophan represses the expression of the trp genes.
- Repression is common in metabolic pathways that result in the biosynthesis of a substance and the co-repressor is normally the end product of the pathway being regulated.
2. Tryptophan operon
a. Structural genes –
- The tryptophan operon (Figure 2.37) contains five structural genes that code for enzymes involved in the synthesis of tryptophan.
- These genes are transcribed from a common promoter into a polycistronic mRNA, which is translated to yield the five enzymes.
b. Regulatory gene –
- The expression of the structural genes is not only influenced by the presence or absence of the co-repressor, it is also controlled by a specific regulatory gene.
- The regulatory gene may be next to or far from the genes that are being regulated. The regulatory gene codes for a specific protein product called a REPRESSOR (sometimes called an apo-repressor).
- When the repressor is synthesized it is inactive. However, it can be activated by complexing with the co-repressor (i.e. tryptophan).
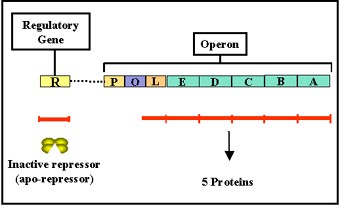
Figure 2.37 : The tryptophan operon
c. Operator –
- The active repressor / co-repressor co-mplex acts by binding to a specific region of the DNA called the operator which is adjacent to the structural genes being regulated.
- The structural genes together with the operator region and the promoter is called an OPERON.
- Thus, in the presence of the co-repressor the repressor is active and binds to the operator, resulting in repression of transcription of the structural genes.
- In contrast, in the absence of co-repressor the repressor is inactive and does not bind to the operator, resulting in transcription of the structural genes.
- This kind of control is referred to a NEGATIVE CONTROL since the function of the regulatory gene product (repressor) is to turn off transcription of the structural genes.
d. Co-repressor –
- Transcription of the tryptophan genes is influenced by the presence or absence of a co-repressor (tryptophan) (Figure 2.38).
- e.g. :- + co-repressor = no expression & – co-repressor = expression
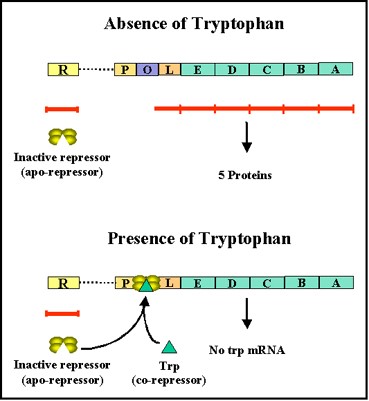
Figure 2.38 : The effect of tryptophan on express ion from the tryptoperon



