RecA & RecBCD
RecA RecBCD
- RecA is a 38 kilodalton Escherichia coli protein essential for the repair and maintenance of DNA.
- RecA has a structural and functional homolog in every species in which it has been seriously sought and serves as an archetype for this class of homologous DNA repair proteins.
- The homologous protein in Homo sapiens is called RAD51.
- RecA has multiple activities, all related to DNA repair.
- In the bacterial SOS response, it has a co-protease function in the autocatalytic cleavage of the LexA repressor and the λ repressor.
- Its most studied role is in facilitating DNA recombination for the repair of double strand DNA breaks and the exchange of genetic information through sexual reproduction.
- E. coli RecA protein also has a major role in homologous recombination. RecA protein binds strongly and in long clusters to ssDNA to form a nucleoprotein filament.
- It has more than one DNA binding site thus RecA can hold a single strand and double strand together.
- This feature makes it possible to catalyze a DNA synapsis reaction between a DNA double helix and a homologous region of single stranded DNA.
- The reaction initiates the exchange of strands between two recombining DNA double helices.
- After the synapis event, in the heteroduplex region a process called branch migration begins.
- In branch migration an unpaired region of one of the single strands displaces a paired region of the other single strand, moving the branch point without changing the total number of base pairs.
- Spontaneous branch migration can occur, however as it generally proceeds equally in both directions it is unlikely to complete recombination efficiently.
- The RecA protein catalyzes unidirectional branch migration and by doing so makes it possible to complete recombination, producing a region of heteroduplex DNA that is thousands of base pairs long.
- RecA protein is a DNA-dependent ATPase, it contains an additional site for binding and hydrolyzing ATP. RecA associates more tightly with DNA when it has ATP bound than when it has ADP bound.
- Escherichia coli strains deficient in RecA are useful for cloning procedures in molecular biology laboratories.
- E. coli strains are often genetically modified to contain a mutant recA locus to ensure the stability of exogenous plasmids: modular circular dsDNA which bacteria replicate with their genome during normal cell growth.
- Plasmid DNA is taken up by the bacteria under a variety of conditions. Bacteria containing exogenous plasmids are called “transformants”.
- Transformants retain the plasmid throughout cell divisions. such that it can be recovered and used in other applications.
- Without functional RecA protein the exogenous plasmid DNA is left unaltered by the bacteria.
- Purification of this plasmid from bacterial cultures then results in high-fidelity amplification of the original plasmid sequence
- The RecA Protein Enables a DNA Single Strand to Pair with a Homologous Region of DNA Double Helix in E. coli 42 General recombination is more complex than the simple hybridization reactions just described.
- In the course of general recombination, a single DNA strand from one DNA double helix must invade another double helix (see Figure 3.8).
fig 3.8
- In E. coli this requires the RecA protein, produced by the recA gene, which was identified in 1965 as being essential for recombination between chromosomes.
- Long sought by biochemists, this elusive gene product was finally purified tohomogeneity in 1976, a feat that allowed its detailed characterization (Figure 3.10).
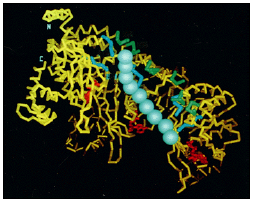
Figure 3.10: The structure of the RecA protein
- Like a singlestrand binding (SSB) protein, the RecA protein binds tightly and in large cooperative clusters to single-stranded DNA to form a nucleoprotein filament.
- This filament has several distinctive properties.
- The RecA protein has more than one DNA-binding site, for example, and it can therefore hold a single strand and a double helix together.
- These sites allow the RecA protein to catalyze a multistep reaction (called synapsis) between a DNA double helix and a homologous region of single-stranded DNA.
- The crucial step in synapsis occurs when a region of homology is identified by an initial base-pairing between complementary nucleotide sequences.
- The nucleation step in this case appears to involve a three-stranded structure, in which the DNA single strand forms nonconventional base pairs in the major groove of the DNA double helix (Figure 3.11).
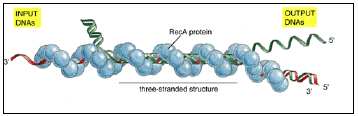
Figure 3.11: DNA synapsis catalyzed by the RecA protein.
- In vitro experiments show that several types of complexes are formed between a DNA single strand covered with RecA protein (red) and a DNA double helix (green).
- First a non-base-paired complex is formed, which is converted to a three-stranded structure as soon as a region of homologous sequence is found.
- This complex is presumably unstable because it involves an unusual form of DNA, and it spins out a DNA heteroduplex (one strand green and the other strand red) plus a displaced single strand from the original helix (green); thus the structure shown in this diagram migrates to the left, reeling in the “input DNAs” while producing the “output DNAs.”
- The net result is a DNA strand exchange identical to that diagrammed earlier in Figure 3.8. (Adapted from S.C. West, Annu. Rev. Biochem. 61:603-640, 1992. © Annual Reviews Inc.)
- This begins the pairing shown previously in Figure 3.8 and so initiates the exchange of strands between two recombining DNA double helices.
- Studies in vitro suggest that the E. coli SSB protein cooperates with the RecA protein to facilitate these reactions.
- Once synapsis has occurred, a short heteroduplex region where the strands from two different DNA molecules have begun to pair is enlarged through protein-directed branch migration, which can also be catalyzed by the RecA protein.
- Branch migration can take place at any point where two single DNA strands with the same sequence are attempting to pair with the same complementary strand; an unpaired region of one of the single strands will displace a paired region of the other single strand, moving the branch point without changing the total number of DNA base pairs.
- Spontaneous branch migration proceeds equally in both directions, and so it makes little progress and is unlikely to complete recombin-ation efficiently (Figure 3.12A).
- Because the RecA protein catalyzes unidirectional branch migration, it readily produces a region of heteroduplex that is thousands of base pairs long (Figure 3.12B).
- The catalysis of branch migration depends on a further property of the RecA protein.
- In addition to having two DNA-binding sites, the RecA protein is a DNA-dependent ATPase, with an additional site for binding and hydrolyzing ATP.
- The protein associates much more tightly with DNA when it has ATP bound than when it has ADP bound.
- Moreover, new RecA molecules with ATP bound are preferentially added at one end of the RecA protein filament, and the ATP is then hydrolyzed to ADP.
- The RecA protein filaments that form on DNA may therefore share many of the dynamic assembly properties displayed by the cytoskeletal filaments formed from actin or tubulin; an ability of the protein to “treadmill” unidirectionally along a DNA strand, for example, could drive the branch migration reaction shown in Figure 3.12B.
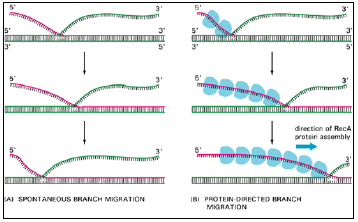
Figure 3.12: Two types of DNA branch migration observed in experiments in vitro.
- (A) Spontaneous branch migration is a back-and-forth, random-walk type of process, and it therefore makes little progress over long distances.
- (B) RecA-protein-directed branch migration proceeds at a uniform rate in one direction, and it may be driven by the polarized assembly of the RecA protein filament on a DNA
RecBCD
- RecBCD, also known as ‘Exonuclease V’, is a protein of the E. coli bacterium that initiates recombinational repair from DNA double strand breaks which are a common result of ionizing radiation, replication errors, endonucleases, oxidative damage and a host of other factors.
- It is both, a helicase that unwinds, or separates the strands of DNA and a nuclease that makes single-stranded nicks in DNA.
- RecBCD (Figure 3.13) is composed of three different subunits, encoded by the recB, recC, and recD genes.
- Both the RecB and RecD subunits are helicases, i.e. energy-dependent molecular motors that unwind DNA or RNA.
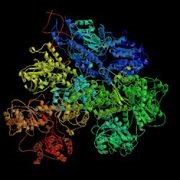
Figure 3.13: RecBCD Crystal Structure
- RecBCD is unusual amongst helicases in that it recognizes a specific sequence in DNA,
- 5′-GCTGGTGG-3′ that is known as ‘Chi’.
- After it initiates unwinding, RecBCD makes nicks on the strand that contains the unwound 3′ end.
- When RecBCD encounters a Chi site on this strand as it is unwinding DNA, it makes a final nick and pauses.
- It has been proposed that this pause is a consequence of a conformational rearrangement in the protein that changes its properties. When RecBCD resumes unwinding, it now nicks the opposite strand (i.e. that containing the 5′ unwound end).
- As a consequence, the 3′ strand remains intact downstream of Chi.
- This is important because the strand exchange protein, RecA, that is responsible for the next step of recombinational repair needs a single-strand molecule with a 3′ end.
- RecBCD is also a model enzyme for the use of single molecule fluorescence as an experimental technique used to better understand the function of protein-DNA interactions.



