MUTATIONS -Spontaneous & Induced Mutations
MUTATIONS -Spontaneous & Induced Mutations
MUTATIONS
- In the living cell, DNA undergoes frequent chemical change, especially when it is being replicated (in S phase of the eukaryotic cell cycle).
- Most of these changes are quickly repaired. Those that are not result in a mutation. Thus, mutation is a failure of DNA repair.
Spontaneous & Induced Mutations
Spontaneous Replication Errors
- Replication is amazingly accurate: fewer than one in a billion errors are made in the course of DNA synthesis.
- However, spontaneous replication errors do occasionally occur.
- The primary cause of spontaneous replication errors was formerly thought to be tautomeric shifts, in which the positions of protons in the DNA bases change.
- Purine and pyrimidine bases exist in different chemical forms called tautomers (Figure 3.21).
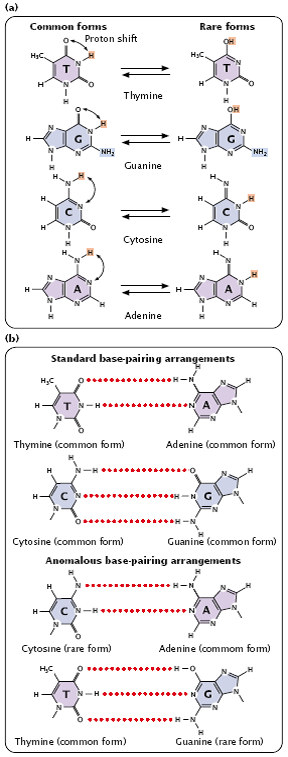
Fig 3.21
- The two tautomeric forms of each base are in dynamic equilibrium, although one form is more common than the other.
- The standard Watson and Crick base pairings—adenine with thymine, and cytosine with guanine—are between the common forms of the bases, but, if the bases are in their rare tautomeric forms, other base pairings are possible (Figure 3.22).
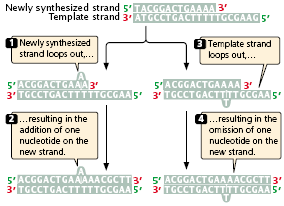
Fig 3.22
- Watson and Crick proposed that tautomeric shifts might produce mutations, and for many years their proposal was the accepted model for spontaneous replication errors, but there has never been convincing evidence that the rare tautomers are the cause of spontaneous mutations.
- Furthermore, research now shows little evidence of these structures in DNA.
- Mispairing can also occur through wobble, in which normal, protonated, and other forms of the bases are able to pair because of flexibility in the DNA helical structure (Figure 3.22).
- These structures have been detected in DNA molecules and are now thought to be responsible for many of the mispairings in replication.
- When a mismatched base has been incorporated into a newly synthesized nucleotide chain, an incorporated error is said to have occurred.
- Suppose that, in replication, thymine (which normally pairs with adenine) mispairs with guanine through wobble (Figure 3.23).
Fig 3.23
- In the next round of replication, the two mismatched bases separate, and each serves as template for the synthesis of a new nucleotide strand.
- This time, thymine pairs with adenine, producing another copy of the original DNA sequence.
- On the other strand, however, the incorrectly incorporated guanine serves as the template and pairs with cytosine, producing a new DNA molecule that has an incorporated error – a C_G pair in place of the original T_A pair (a T_A:C_G base substitution).
- The original incorporated error leads to a replication error, which creates a permanent mutation, because all the base pairings are correct and there is no mechanism for repair systems to detect the error.
- Mutations due to small insertions and deletions also may arise spontaneously in replication and crossing over.
- Strand slippage may occur when one nucleotide strand forms a small loop (Figure 3.24).

- If the looped-out nucleotides are on the newly synthesized strand, an insertion results.
- At the next round of replication, the insertion will be incorporated into both strands of the DNA molecule.
- If the looped-out nucleotides are on the template strand, then there is a deletion on the newly replicated strand, and this deletion will be perpetuated in subsequent rounds of replication.
- During normal crossing over, the homologous sequences of the two DNA molecules align, and crossing over produces no net change in the number of nucleotides in either molecule.
- Misaligned pairing may cause unequal crossing over, which results in one DNA molecule with an insertion and the other with a deletion (Figure 3.25).
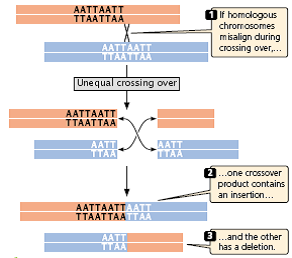
Fig 3.25
- Some DNA sequences are more likely than others to undergo strand slippage or un-equal crossing over. Stretches of repeated sequences, such as trinucleotide repeats or homopolymeric repeats (more than five repeats of the same base in a row), are prone to strand slippage.
- Stretches with more repeats are more likely to undergo strand slippage.
- Duplicated or repetitive sequences may misalign during pairing, leading to unequal crossing over.
- Both strand slippage and unequal crossing over produce duplicated copies of sequences, which in turn promote further strand slippage and unequal crossing over.
- This chain of events may explain the phenomenon of anticipation often observed for expanding trinucleotide repeats.
Spontaneous Chemical Changes –
- In addition to spontaneous mutations that arise in replication, mutations also result from spontaneous chemical changes in DNA.
- One such change is depurination, the loss of a purine base from a nucleotide.
- Depurination results when the covalent bond connecting the purine to the 1_-carbon atom of the deoxyribose sugar breaks (Figure 3.26), producing an apurinic site – a nucleotide that lacks its purine base.
- An apurinic site cannot act as a template for a complementary base in replication.
Fig 3.26
- In the absence of base-pairing constraints, an incorrect nucleotide (most often adenine) is incorporated into the newly synthesized DNA strand opposite the apurinic site (Figure 3.26b), frequently leading to an incorporated error.
- The incorporated error is then transformed into a replication error at the next round of replication.
- Depurination is a common cause of spontaneous mutation; a mammalian cell in culture loses approximately 10,000 purines every day.
- Another spontaneously occurring chemical change that takes place in DNA is deamination, the loss of an amino group (NH2) from a base.
- Deamination may occur spontaneously or be induced by mutagenic chemicals
Chemically Induced Mutations –
- Although many mutations arise spontaneously, a number of environmental agents are capable of damaging DNA, including certain chemicals and radiation.
- Any environmental agent that significantly increases the rate of mutation above the spontaneous rate is called a mutagen.
(1) Base analogs –
- one class of chemical mutagens consists of base analogs, chemicals with structures similar to that of any of the four standard bases of DNA. DNA polymerases cannot distinguish these analogs from the standard bases; so,
- if base analogs are present during replication, they may be incorporated into newly synthesized DNA molecules.
- For example, 5-bromouracil (5BU) is an analog of thymine; it has the same structure as that of thymine except that it has a bromine (Br) atom on the 5-carbon atom instead of a methyl group (Figure 3.28a).
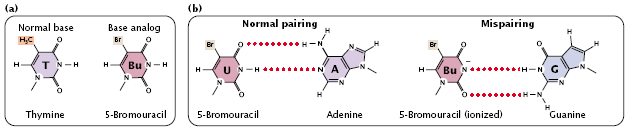
fig 3.28
- Normally, 5-bromouracil pairs with adenine just as thymine does, but it occasionally mispairs with guanine (Figure 3.28b), leading to a transition (T_A:5BU_A:5BU_G:C_G), as shown in (Figure 3.31).
- Through mispairing, 5-bromouracil may also be incorporated into a newly synthesized DNA strand opposite guanine. In the next round of replication, 5-bromouracil may pair with adenine, leading to another transition (G_C:G_5BU:A_5BU:A_T).
(2) 2-aminopurine (2AP) –
- Another mutagenic chemical is, which is a base analog of adenine.
- Normally, 2-aminopurine base pairs with thymine, but it may mispair with cytosine, causing a transition mutation (T_A:T_2AP:C_2AP:C_G).
- Alternatively, 2-aminopurine may be incorporated through mispairing into the newly synthesized DNA opposite cytosine and later pair with thymine, leading to a C_G:C_2AP:T_2AP:T_A transition.
- Thus, both 5-bromouracil and 2-aminopurine can produce transition mutations. In the laboratory, mutations by base analogs can be reversed by treatment with the same analog or by treatment with a different analog.
(3) Alkylating agents –
- Alkylating agents are chemicals that donate alkyl groups.
- These agents include methyl (CH3) and ethyl (CH3–CH2) groups, which are added to nucleotide bases by some chemicals.
- For example, ethylmethanesulfonate (EMS) adds an ethyl group to guanine, producing 6-ethylguanine, which pairs with thymine (see Figure 3.29a).
- Thus, EMS produces C_G:T_A transitions. EMS is also capable of adding an ethyl group to thymine, producing 4-ethylthymine, which then pairs with guanine, leading to a T_A:C_G transition.
- Because EMS produces both C_G:T_A and T_A:C_G transitions, mutations produced by EMS can be reversed by additional treatment with EMS. Mustard gas is another alkylating agent.
(4) Deamination –
- In addition to its spontaneous occurrence (see Figure 3.27), deamination can be induced by some chemicals.
- For instance, nitrous acid deaminates cytosine, creating uracil, which in the next round of replication pairs with adenine (see Figure 3.29b), producing a C_G:T_A transition mutation.
- Nitrous acid changes adenine into hypoxanthine, which pairs with cytosine, leading to a T_A:C_G transition.
- Nitrous acid also deaminates guanine, producing xanthine, which pairs with cytosine just as guanine does; however xanthine may also pair with thymine, leading to a C_G:T_A transition.
- Nitrous acid produces exclusively transition mutations and, because both C_G:T_A and T_A:C_G transitions are produced, these mutations can be reversed with nitrous acid.
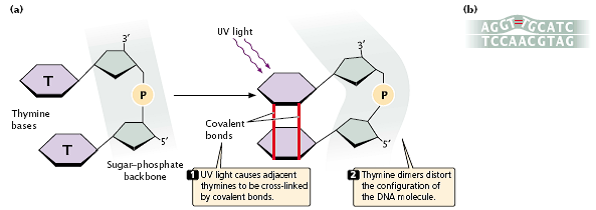
fig 3.27
- Hydroxylamine Hydroxylamine is a very specific basemodifying mutagen that adds a hydroxyl group to cytosine, converting it into hydroxylaminocytosine.
- This conversion increases the frequency of a rare tautomer that pairs with adenine instead of guanine and leads to C_G:T_A transitions.
- Because hydroxylamine acts only on cytosine, it will not generate T_A:C_G transitions; thus, hydroxylamine will not reverse the mutations that it produces.
(5) Oxidative reactions –
- Reactive forms of oxygen (including superoxide radicals, hydrogen peroxide, and hydroxyl radicals) are produced in the course of normal aerobic metabolism, as well as by radiation, ozone, peroxides, and certain drugs.
- These reactive forms of oxygen damage DNA and induce mutations by bringing about chemical changes to DNA.
- For example, oxidation converts guanine into 8-oxy-7,8-dihydrodeoxyguanine (Figure 3.30), which frequently mispairs with adenine instead of cytosine, causing a G_C:T_A transversion mutation.
Fig 3.30
(6) Intercalating agents –
- Intercalating agents, such as proflavin, acridine orange, ethidium bromide, and dioxin are about the same size as a nucleotide (Figure 3.32a).
- They produce mutations by sandwiching themselves (intercalating) between adjacent bases in DNA, distorting the three-dimensional structure of the helix and causing single- nucleotide insertions and deletions in replication (Figure 3.32b).
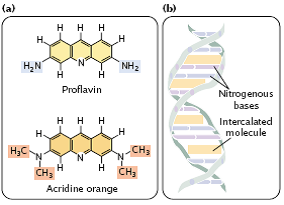
fig 3.32
- These insertions and deletions frequently produce frameshift mutations (which change all amino acids downstream of the mutation), and so the mutagenic effects of intercalating agents are often severe.
- Because intercalating agents generate both additions and deletions, they can reverse the effects of their own mutations.
Radiation –
- In 1927, Herman Muller demonstrated that mutations in fruit flies could be induced by X-rays.
- The results of subsequent studies showed that X-rays greatly increase mutation rates in all organisms.
- The high energies of X-rays, gamma rays, and cosmic rays (Figure 3.33) are all capable of penetrating tissues and damaging DNA.
- These forms of radiation, called ionizing radiation, dislodge electrons from the atoms that they encounter, changing stable molecules into free radicals and reactive ions, which then alter the structures of bases and break phosphodiester bonds in DNA.
- Ionizing radiation also frequently results in double-strand breaks in DNA. Attempts to repair these breaks can produce chromosome mutations.

Fig 3.33
- Ultraviolet light has less energy than that of ionizing radiation and does not eject electrons and cause ionization but is nevertheless highly mutagenic.
- Purine and pyrimidine bases readily absorb UV light, resulting in the formation of chemical bonds between adjacent pyrimidine molecules on the same strand of DNA and in the creation of structures called pyrimidine dimmers (Figure 3.34a).
- Pyrimidine dimers consisting of two thymine bases (called thymine dimers) are most frequent, but cytosine dimmers and thymine–cytosine dimers also can form.
- These dimmers distort the configuration of DNA (Figure 3.34b) and often block replication.

fig 3.34
- Most pyrimidine dimers are immediately repaired by mechanisms discussed later in this chapter, but some escape repair and inhibit replication and transcription.
- When pyrimidine dimers block replication, cell division is inhibited and the cell usually dies; for this reason, UV light kills bacteria and is an effective sterilizing agent.
- For a mutation—a hereditary error in the genetic instructions – to occur, the replication block must be overcome.
- How do bacteria and other organisms replicate despite the presence of thymine dimers? Bacteria can circumvent replication blocks produced by pyrimidine dimers and other types of DNA damage by means of the SOS system.
- This system allows replication blocks to be overcome, but in the process makes numerous mistakes and greatly increases the rate of mutation. Indeed, the very reason that replication can proceed in the presence of a block is that the enzymes in the SOS system do not strictly adhere to the base-pairing rules.
- The trade-off is that replication may continue and the cell survives, but only by sacrificing the normal accuracy of DNA synthesis.
- The SOS system is complex, including the products of at least 25 genes.
- A protein called RecA binds to the damaged DNA at the blocked replication fork and becomes activated.
- This activation promotes the binding of a protein called LexA, which is a repressor of the SOS system.
- The activated RecA complex induces LexA to undergo self-cleavage, destroying its repressive activity.
- This inactivation enables other SOS genes to be expressed, and the products of these genes allow replication of the damaged DNA to proceed.
- The SOS system allows bases to be inserted into a new DNA strand in the absence of bases on the template strand, but these insertions result in numerous errors in the base sequence.
- Eukaryotic cells have a specialized DNA polymerase called polymerase η (eta) that bypasses pyrimidine dimers.
- Polymerase η preferentially inserts AA opposite a pyrimidine dimer.
- This strategy seems to be reasonable because about two-thirds of pyrimidine dimers are thymine dimers.
- However, the insertion of AA opposite a CT dimer results in a C_G:A_T transversion. Polymerase η is therefore said to be an error-prone polymerase.



