Molecular Genetic Map
These maps are prepared utilizing the following molecular markers:
I. Non-PCR Based:
(a) Restriction Fragment Length Polymorphism (RFLP):
- When the genomic DNA from several individuals (genetically related) are digested with same restriction enzymes and separated on gel through electrophoresis, blotted and hybridized with a labelled DNA clone of known sequence, polymorphism in hybridization pattern reveals the relatedness among different individuals, and such variation is termed as ‘Restriction Fragment Length Polymorphism’ (RFLP) (Fig 22.15).
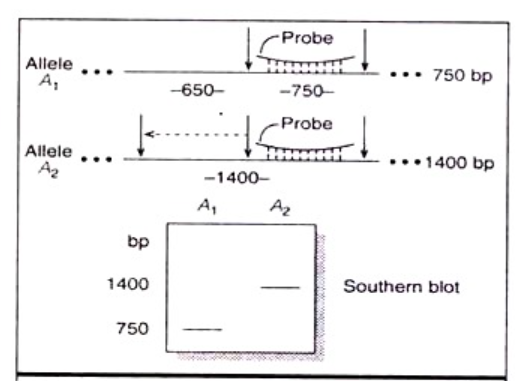
- RFLP is used to infer phylogenetic relationship in both plants and animals.
- This technique is also used to prepare chromosome maps in human, mice, fruitfly, or in plants like maize, tomato, lettuce and rice.
- The inheritance or linkage relationship also can be studied through this technique analysing the parent, F1 and F2 population.
II. PCR based:
(b) Random Amplified Polymorphic DNA (RAPD):
- If artificially synthesized primers with arbitrary sequences are used for PCR (Polymerase chain reaction) amplification,
- DNA segments to be amplified will be selected at random which will thus provide a truly random sample of DNA markers, which are described as ‘Random Amplified Polymorphic DNA’ (RAPD).
The steps involved are:
(i) Amplification of extracted DNA by PCR using random primers.
(ii) Separation of amplified DNA on agarose gel.
(iii) Visualization of markers on the gel and its photography.
(c) Amplified Fragment Length Polymorphism (AFLP):
- This technique involves the ligation of adaptors to DNA fragments obtained after RE digestion. Then these fragments are allowed for PCR.
- The primers used here are generally sequence of adaptors which help to amplify the whole length between the two restriction sites.
- Then the different fragment lengths are separated on gel.
(d) SCAR (Sequence Characterised Amplified Region):
- Amplified RAPD products are cloned, sequenced and primer sequences are ascertained from the end of band, the primers are generally longer which gives greater degree of specificity.
(e) Variable Number Tandem Repeats (VNTR):
- The DNA differences between two individuals can be detected by variable number of tandem repeats which are short repeat sequences arranged in tandem order.
- VNTRs are divided into two groups Minisatellites (10-25 bps) and Microsatellites (1-5 bps).
- A DNA poly-core probe of such micro- or minisatellites can detect simultaneously a large number of variable loci containing tandem DNA repeat sequences.
(f) Sequence-Tagged Sites (STS):
- Sequence tagged sites are DNA length of 100-500 bp that are unique in genome.
- They are created by polymerase chain reaction amplification of primers obtained by sequencing segments of the genome.
(g) Single Nucleotide Polymorphism (SNP):
- The map can also be marked off in differences among individuals that amount to changes in single base pairs. These differences are called SNPs.