Mitochondrial DNA Replication
Mitochondrial DNA (mtDNA) replication is a unique process distinct from nuclear DNA replication. Unlike nuclear DNA, which replicates in the S-phase of the cell cycle, mtDNA replicates independently throughout the cell cycle.
Features of mtDNA Replication
- Independent Replication – Unlike nuclear DNA, mtDNA replication is not synchronized with the cell cycle.
- Circular DNA – mtDNA is a double-stranded, circular DNA molecule similar to bacterial DNA.
- Unidirectional & Asynchronous – The replication occurs in a strand-displacement manner rather than a traditional bidirectional mode seen in nuclear DNA.
- Two Origin Sites
- OH (Origin of Heavy strand synthesis) – Replication of the heavy strand (rich in G nucleotides).
- OL (Origin of Light strand synthesis) – Replication of the light strand (rich in C nucleotides).
- No Histone Proteins – Unlike nuclear DNA, mtDNA is not associated with histones, making it more susceptible to mutations.
- Enzymes Involved – mtDNA replication uses specialized polymerases and enzymes different from nuclear DNA replication.
Steps of Mitochondrial DNA Replication
- Initiation
- The replication starts at OH (Origin of Heavy strand) in the D-loop region.
- mtDNA helicase (Twinkle) unwinds the double strand.
- Mitochondrial single-strand binding protein (mtSSB) stabilizes the unwound strand.
- Elongation
- DNA Polymerase γ (gamma) synthesizes the heavy strand using the light strand as a template.
- As the heavy strand elongates, OL (Origin of Light strand synthesis) is exposed.
- The light strand synthesis begins using a separate primer.
- Termination
- Replication continues until both strands are completely synthesized.
- The daughter molecules separate, and mtDNA is ready for transcription and translation.
Enzymes Involved in mtDNA Replication
| Enzyme | Function |
|---|---|
| DNA Polymerase γ (POLG) | Main enzyme for mtDNA replication |
| Twinkle Helicase | Unwinds the DNA strands |
| mtSSB (Mitochondrial Single-Strand Binding Protein) | Prevents strand reannealing |
| Topoisomerase | Relieves supercoiling |
| RNase H1 | Removes RNA primers |
| DNA Ligase III | Seals the nicks in the DNA strands |
Mitochondrial DNA Replication Models
Mitochondrial DNA (mtDNA) replication differs from nuclear DNA replication in several ways, mainly because mtDNA is circular, lacks histones, and replicates independently of the cell cycle. Various models have been proposed to explain how mtDNA replication occurs.
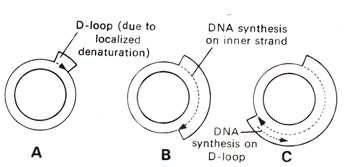
1. Strand-Displacement Model (Accepted Model)
The strand-displacement model is the most widely accepted model for animal mitochondrial DNA replication.
Features
✅ Unidirectional replication – Heavy and light strands are replicated separately.
✅ Asynchronous synthesis – The heavy strand starts replication first, followed by the light strand.
✅ D-loop formation – A triple-stranded structure is formed temporarily.
Mechanism:
- Initiation:
- Replication begins at the Origin of Heavy strand (OH), which is located within the D-loop region of mtDNA.
- The Twinkle helicase unwinds the DNA, and mtSSB (Mitochondrial Single-Strand Binding Protein) stabilizes the single strand.
- DNA Polymerase γ (POLG) starts synthesizing the new heavy strand, displacing the original heavy strand.
- Strand Displacement:
- As the heavy strand elongates, it displaces the original heavy strand, forming a D-loop structure.
- Once the Origin of Light strand (OL) is exposed, replication of the light strand begins.
- DNA Polymerase γ synthesizes the light strand in the opposite direction.
- Completion:
- Both strands complete synthesis independently.
- The D-loop resolves, forming two circular mtDNA molecules.
Significance:
🔹 Found in mammals, including humans.
🔹 Explains the presence of D-loops in mtDNA.
🔹 Efficient for high-energy-demanding cells.
2. Synchronous Model (Alternative Model)
The synchronous model, also known as the strand-coupled bidirectional model, proposes that both strands are synthesized simultaneously.
Features
✅ Bidirectional replication – Similar to nuclear DNA replication.
✅ Synchronous strand synthesis – Both heavy and light strands are synthesized together.
✅ Two replication forks move in opposite directions.
Mechanism
- Replication starts at a single origin (OriH/OriL) and proceeds in both directions.
- Two replication forks move away from the origin, synthesizing both strands at the same time.
- DNA polymerase γ replicates both strands continuously (leading strand) and discontinuously (lagging strand).
- The replication forks meet on the opposite side, completing the synthesis.
Significance
🔹 Found in yeast, fungi, and some protists.
🔹 More similar to bacterial DNA replication.
🔹 Less evidence in higher eukaryotes like mammals.
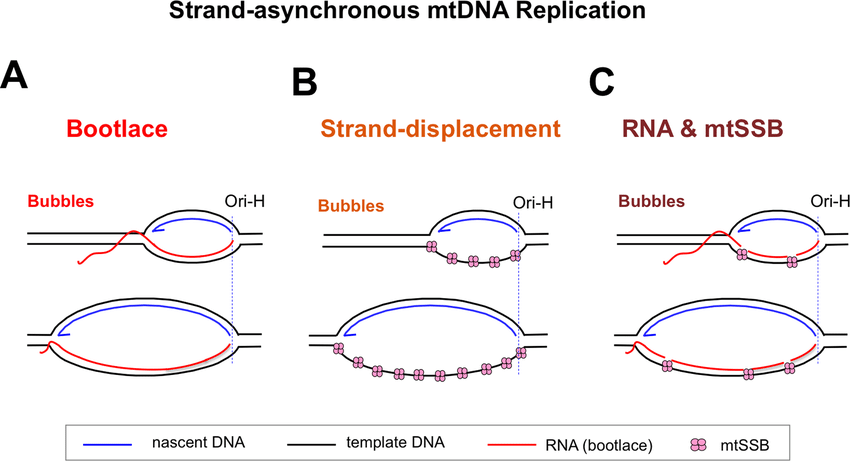
3. RITOLS Model (RNA Incorporation Throughout the Lagging Strand)
The RITOLS (RNA Incorporation Throughout the Lagging Strand) model suggests that RNA is used as a primer and template during replication.
Key Features
✅ RNA intermediates replace DNA in the lagging strand.
✅ Gradual replacement of RNA by DNA.
✅ Strand-displacement mechanism is still involved.
Mechanism
- The heavy strand initiates replication, and the lagging strand is temporarily made of RNA instead of DNA.
- The RNA is then gradually replaced by DNA through RNase H and DNA polymerase γ.
- The final mtDNA molecule contains only double-stranded DNA.
Significance
🔹 Explains some observed RNA intermediates in mitochondria.
🔹 Found in mammals and some vertebrates.
🔹 Still under investigation.
4. Recombination-Dependent Model
This model suggests that mtDNA replication involves recombination events, similar to nuclear DNA.
Features
✅ Homologous recombination plays a role in mtDNA replication.
✅ Replication forks can be restarted after stalling.
✅ More common in plants and lower eukaryotes.
Mechanism
- Replication forks stall due to DNA damage or stress.
- Homologous recombination repairs the stalled fork.
- Replication resumes after recombination-mediated repair.
Significance
🔹 Found in plants, yeast, and some protists.
🔹 May help in mtDNA repair and maintenance.

Comparison of Mitochondrial DNA Replication Models
| Model | Strand Displacement | Synchronous | RITOLS | Recombination-Dependent |
|---|---|---|---|---|
| Replication Type | Unidirectional | Bidirectional | Unidirectional | Bidirectional |
| Strand Synthesis | Asynchronous | Synchronous | Asynchronous | Asynchronous |
| RNA Involvement | Low | None | High | Moderate |
| D-Loop Formation | Yes | No | Yes | No |
| Main Organisms | Mammals | Yeast, Fungi | Mammals | Plants, Yeast |
Multiple-Choice Questions (MCQs) on Mitochondrial DNA Replication
1. Which model is most widely accepted for mitochondrial DNA replication in mammals?
A) Strand-Displacement Model
B) Synchronous Model
C) RITOLS Model
D) Recombination-Dependent Model
Answer: A) Strand-Displacement Model
2. What is the role of DNA Polymerase γ in mitochondrial DNA replication?
A) It helps in the unwinding of DNA
B) It is responsible for mtDNA transcription
C) It synthesizes new mitochondrial DNA strands
D) It repairs damaged mitochondrial DNA
Answer: C) It synthesizes new mitochondrial DNA strands
3. In the strand-displacement model, where does replication of the heavy strand begin?
A) Origin of Replication in Nucleus
B) D-Loop Region
C) Cytoplasm
D) Mitochondrial Outer Membrane
Answer: B) D-Loop Region
4. Which enzyme is responsible for unwinding the mitochondrial DNA during replication?
A) DNA Polymerase γ
B) Twinkle Helicase
C) Topoisomerase
D) RNase H
Answer: B) Twinkle Helicase
5. What type of DNA structure is present in mitochondria?
A) Linear double-stranded DNA
B) Circular double-stranded DNA
C) Single-stranded DNA
D) Supercoiled RNA
Answer: B) Circular double-stranded DNA
6. What distinguishes mitochondrial DNA replication from nuclear DNA replication?
A) It occurs only in the S phase
B) It is independent of the cell cycle
C) It uses DNA Polymerase α
D) It has only one replication fork
Answer: B) It is independent of the cell cycle
7. What is the function of RNase H in the RITOLS model?
A) It synthesizes the heavy strand
B) It removes RNA primers and replaces them with DNA
C) It unwinds mitochondrial DNA
D) It synthesizes ATP
Answer: B) It removes RNA primers and replaces them with DNA
8. Which mitochondrial DNA replication model suggests the presence of RNA intermediates in the lagging strand?
A) Strand-Displacement Model
B) Synchronous Model
C) RITOLS Model
D) Recombination-Dependent Model
Answer: C) RITOLS Model
9. Which protein regulates both mitochondrial DNA replication and transcription?
A) DNA Polymerase γ
B) TFAM (Mitochondrial Transcription Factor A)
C) Twinkle Helicase
D) ATP Synthase
Answer: B) TFAM (Mitochondrial Transcription Factor A)
10. What is the primary function of mitochondrial DNA?
A) To encode all proteins in the mitochondria
B) To store genetic information for the entire cell
C) To encode proteins, tRNA, and rRNA required for mitochondrial function
D) To regulate nuclear DNA transcription
Answer: C) To encode proteins, tRNA, and rRNA required for mitochondrial function
11. The light strand of mitochondrial DNA begins replication at which site?
A) Origin of Heavy Strand (OH)
B) Origin of Light Strand (OL)
C) D-Loop
D) Mitochondrial Membrane
Answer: B) Origin of Light Strand (OL)
12. Which of the following is a disorder caused by mitochondrial DNA replication defects?
A) Huntington’s Disease
B) Kearns-Sayre Syndrome (KSS)
C) Alzheimer’s Disease
D) Down Syndrome
Answer: B) Kearns-Sayre Syndrome (KSS)
13. Which organism has the smallest mitochondrial genome?
A) Humans
B) Yeast
C) Plasmodium falciparum
D) Plants
Answer: C) Plasmodium falciparum
14. How does ethidium bromide affect mitochondrial DNA replication?
A) It enhances mtDNA replication
B) It inhibits mtDNA replication and causes deletions
C) It acts as a mitochondrial DNA polymerase
D) It increases ATP production
Answer: B) It inhibits mtDNA replication and causes deletions
15. In which phase of the cell cycle does mitochondrial DNA replication primarily occur?
A) S Phase
B) G1 Phase
C) It occurs continuously, independent of the cell cycle
D) M Phase
Answer: C) It occurs continuously, independent of the cell cycle
16. Which enzyme is responsible for maintaining mitochondrial DNA integrity by proofreading errors during replication?
A) DNA Polymerase γ
B) RNA Polymerase
C) Helicase
D) Reverse Transcriptase
Answer: A) DNA Polymerase γ
17. The origin of replication for the heavy strand (OH) is located within which mitochondrial structure?
A) Inner mitochondrial membrane
B) Outer mitochondrial membrane
C) D-Loop
D) Cristae
Answer: C) D-Loop
18. Which mitochondrial DNA replication model suggests bidirectional replication?
A) Strand-Displacement Model
B) Synchronous Model
C) RITOLS Model
D) Recombination-Dependent Model
Answer: B) Synchronous Model
19. What is a unique feature of mitochondrial DNA compared to nuclear DNA?
A) Presence of histones
B) Linear structure
C) Maternal inheritance
D) Larger genome size
Answer: C) Maternal inheritance
20. Which protein helps unwind mitochondrial DNA during replication?
A) Helicase
B) DNA Ligase
C) Primase
D) Gyrase
Answer: A) Helicase
21. What happens to mitochondrial DNA during mitophagy?
A) It undergoes excessive replication
B) It is selectively degraded
C) It moves to the nucleus
D) It forms a new mitochondrial network
Answer: B) It is selectively degraded
22. The majority of proteins required for mitochondrial DNA replication are encoded by which genome?
A) Mitochondrial genome
B) Chloroplast genome
C) Nuclear genome
D) Bacterial genome
Answer: C) Nuclear genome
23. Which of the following statements about mitochondrial DNA is FALSE?
A) It lacks histones
B) It is inherited maternally
C) It contains introns
D) It codes for essential mitochondrial proteins
Answer: C) It contains introns
24. Which enzyme is responsible for the removal of RNA primers during mitochondrial DNA replication?
A) DNA Polymerase γ
B) RNase H
C) Ligase
D) Helicase
Answer: B) RNase H
25. How is mitochondrial DNA replication different from nuclear DNA replication?
A) It follows the semi-conservative model
B) It uses histone proteins
C) It occurs throughout the cell cycle
D) It has telomeres
Answer: C) It occurs throughout the cell cycle
26. What is the role of TFAM (Mitochondrial Transcription Factor A) in mtDNA replication?
A) It acts as an ATP synthase
B) It helps regulate mtDNA replication and transcription
C) It degrades old mitochondrial DNA
D) It functions in glycolysis
Answer: B) It helps regulate mtDNA replication and transcription
27. What does the mitochondrial DNA D-Loop contain?
A) Only non-coding regions
B) Only protein-coding genes
C) The replication origin for the heavy strand
D) The enzyme ATP Synthase
Answer: C) The replication origin for the heavy strand
28. What is the size of the human mitochondrial genome?
A) 500 bp
B) 16,579 bp
C) 100,000 bp
D) 3 million bp
Answer: B) 16,579 bp
29. What is the significance of the petite mutation in yeast mitochondrial DNA?
A) It enhances mitochondrial function
B) It leads to defective mitochondrial function
C) It increases mitochondrial genome size
D) It promotes nuclear DNA replication
Answer: B) It leads to defective mitochondrial function
30. Which enzyme is required for mitochondrial DNA ligation during replication?
A) DNA Helicase
B) DNA Ligase III
C) Topoisomerase
D) Primase
Answer: B) DNA Ligase III
31. How is mitochondrial DNA inherited in most organisms?
A) Paternally
B) Biparentally
C) Maternally
D) Randomly
Answer: C) Maternally
32. What happens when mitochondrial DNA replication is defective?
A) Increased ATP production
B) Mitochondrial dysfunction and disease
C) Nuclear DNA replication failure
D) Enhanced oxygen consumption
Answer: B) Mitochondrial dysfunction and disease
33. Which mitochondrial DNA replication model involves strand-coupled replication?
A) Synchronous Model
B) Strand-Displacement Model
C) RITOLS Model
D) End-Replication Model
Answer: A) Synchronous Model
34. What is the primary function of mitochondrial DNA?
A) Encode all mitochondrial proteins
B) Encode proteins involved in oxidative phosphorylation
C) Replicate nuclear DNA
D) Regulate gene expression in the nucleus
Answer: B) Encode proteins involved in oxidative phosphorylation
35. What is the role of mitochondrial ribosomes (55S or 70S)?
A) Degrade mitochondrial DNA
B) Synthesize mitochondrial proteins
C) Regulate ATP production
D) Control mitochondrial division
Answer: B) Synthesize mitochondrial proteins
36. Which enzyme removes supercoils in mitochondrial DNA?
A) DNA Ligase III
B) Topoisomerase
C) DNA Polymerase γ
D) RNase H
Answer: B) Topoisomerase
37. Which process ensures that damaged mitochondrial DNA is eliminated?
A) Mitosis
B) Mitophagy
C) Glycolysis
D) Transcription
Answer: B) Mitophagy
38. What is the significance of the RITOLS model of mitochondrial DNA replication?
A) It proposes that replication occurs in the absence of RNA
B) It suggests an RNA component in the lagging strand
C) It denies the existence of mitochondrial DNA
D) It explains why mitochondria lack ribosomes
Answer: B) It suggests an RNA component in the lagging strand
39. What happens when mitochondrial DNA accumulates mutations?
A) Cells increase ATP production
B) Cells develop mitochondrial diseases
C) Cells grow faster
D) The mitochondrial genome expands
Answer: B) Cells develop mitochondrial diseases
40. What is the function of Twinkle helicase?
A) It transcribes mitochondrial DNA
B) It unwinds mitochondrial DNA during replication
C) It degrades ribosomal RNA
D) It attaches mitochondria to the nucleus
Answer: B) It unwinds mitochondrial DNA during replication
41. Which enzyme synthesizes RNA primers for mitochondrial DNA replication?
A) Primase
B) DNA Ligase III
C) ATP Synthase
D) Ubiquinone
Answer: A) Primase
42. How does mitochondrial DNA replication contribute to aging?
A) It repairs telomeres
B) Mutations in mtDNA accumulate over time, leading to cell dysfunction
C) It increases nuclear DNA stability
D) It stops after a certain age
Answer: B) Mutations in mtDNA accumulate over time, leading to cell dysfunction
43. What is a common feature of mitochondrial diseases?
A) Defects in oxidative phosphorylation
B) Increased mitochondrial DNA replication
C) Complete loss of nuclear DNA
D) Resistance to antibiotics
Answer: A) Defects in oxidative phosphorylation
