Construction of Genomic and cDNA Library
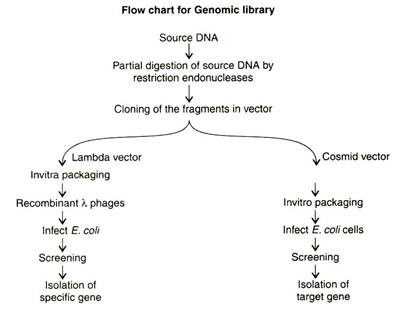
- A genomic library contains all the sequences present in the genome of an organism. In the construction of genomic libraries it is feasible to use vectors that could accommodate large sizes of inserts.
- The first step in the construction of a genomic library is the isolation of genomic DNA, and entire DNA is subjected to restriction digestion.
- The fragmented DNA of suitable size is ligated in the appropriate cloning vectors.
- It is necessary to use partially digested DNA with partially used restriction enzymes to generate a random collection of fragments with a suitable size distribution.
- The recombination vectors are transferred and maintained in organisms such as bacteria, viruses or yeast.
- A target DNA sequence present in particular cell clones are identified, sub-cultured and maintained as cell lines, widely known as gene bank or a clone bank.
Steps in Genomic Library Construction:
(а) Isolation of target DNA:
- Genomic libraries can be constructed by isolation of complete DNA from bacteria, virus, plants and animals.
- In eukaryotes, high molecular weight DNA is isolated by CTAB or SDS methods.
- The isolated DNA is then purified by caesium chloride and other methods.
(b) Restriction Fragments:
- Fragmentation can be done by mechanical shearing or using suitable restriction enzymes.
- Partial digestion is essential to procure proper size DNA fragments.
- Therefore, treatment times and concentration of enzymes are very important for desirable results.
(c) Cloning the fragments in vector:
- The restricted digested DNA sample is electrophoresed and subjected to. Target DNA fragments are identified by hybridization with probes and then cloned in suitable vectors like lambda or cosmid vectors and maintained as a library.
(d) Screening of Genomic library:
- Genomic libraries can be screened for clones by hybridization with probe, western blotting to detect protein product and also screening of protein activity.
Screening by Colony Hybridization:
- Principally this screening technique involves hybridization between labelled DNA probes and a target DNA sequence.
- Therefore, a target gene sequence can be accurately identified.
The essential steps are described as follows:
- A colony of host cells is plated onto a solid medium containing antibiotics.
- The presence of antibiotics in the medium ensures growth of only transformed cells due to its antibiotic resistance gene in their plasmid.
- When a discrete colony is formed on the plate, it is then transferred onto nitrocellulose or nylon membrane commonly referred as solid matrix.
- The exact position of the cell colony on the plate is maintained on the matrix.
- Once the cells are attached to solid matrix (nitrocellulose paper) cells are lysed, deproteinized and released DNA is denatured by alkaline reagent.
- The labelled DNA probe is mixed in the next step, to facilitate hybridization between target DNA and DNA probe. The unbound probe is washed off from the matrix
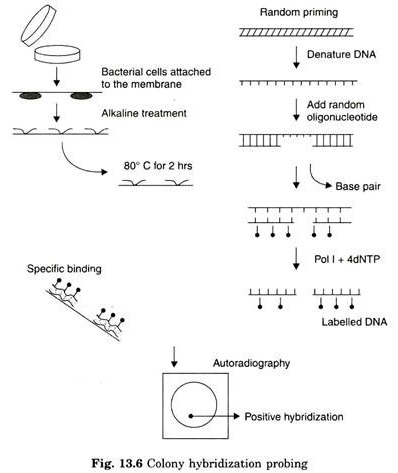
- Matrix is then subjected to autoradiography to determine the location of cells containing hybridized DNA.
- Development of the dark spot on the X-ray film indicates the presence of target DNA (gene).
- The dark spot on X-ray film corresponds to the cell colony on the master plate.
- Once a transformed cell colony is identified, it is then sub-cultured and maintained as it carries cloned DNA of interest.
Plaque Hybridization:
- A genomic library is screened by infecting phage viruses containing inserted DNA on a lawn of bacteria.
The steps are described as follows:
- A lawn of bacterial cultures is initiated on the agar plate.
- Several thousands of phage particles containing inserted DNA are spread on the lawn of bacteria.
- Phage infects bacteria and plaques are obtained.
- These are then transferred to nitrocellulose paper; DNA is denatured to single strand and bound firmly onto solid matrix.
- 32P labelled DNA/RNA probe is added to ensure hybridization.
- The location of the bound probe is determined by hybridization.
- The position of the spot on the X-ray film corresponds to the original site on a agar plate.
- Once the desired gene is identified, it is then isolated and sub-cultured.
Immunological Screening:
- This is another novel method of screening a gene library.
- This method can be used as an alternative to DNA probes.
- The cells are allowed to transcribe and translate and the presence of protein can be identified by corresponding antibody molecules.
The technique is described as follows:
- Discrete colonies are formed on the master plate,
- Transfer sample of cells from each colony to nitrocellulose or nylon membrane.
- Allow the cells to transcribe and translate.
- The cells are then lysed and proteins are bound to the matrix.
- Initially primary antibody treatment is done to facilitate the binding of primary antibody to protein molecules bound on the solid matrix.
- Add a secondary antibody that binds only to the primary antibody.
- Colorimetric reaction takes place only if the secondary antibody is present (Enzyme bound secondary antibody develops coloured compound when treated with respective substrate).
- A colony on the master plate that corresponds to a positive response on the matrix is identified.
- The cells from the positive colony on the master plate are sub-cultured as this may carry insert DNA that encodes the protein that binds the primary antibody.
Chemiluminescence:
- Chemiluminescence is a process that emits light due to chemical reaction.
- The chemiluminescent reactions are catalyzed by enzymes such as horseradish peroxidase (HRP) and alkaline phosphatase (AP).
- In this reaction, enzyme substrates are converted to products, releasing photons of a defined wavelength.
- For example, dioxetanes react with peroxides in the presence of HRP to emit a flash of blue light of about 435 nm.
- Direct detection methods use direct covalent attachment of the signalling enzyme to the nucleic acid probe like synthetic oligonucleotides.