![]()
Molecular Marker
Molecular Marker for Introgression of useful traits
- In genetics, a molecular marker(identified as genetic marker) is a fragment of DNA that is associated with a certain location within the genome. Molecular markers are used in molecular biology and biotechnology to identify a particular sequence of DNA in a pool of unknown DNA.
- Molecular markers are nucleotide sequences and can be investigated through the polymorphism present between the nucleotide sequences of different individuals.
- Insertion, deletion, point mutations duplication and translocation are basis of these polymorphisms; however, they do not necessarily affect the activity of genes.
- An ideal DNA marker should be co-dominant, evenly distributed throughout genome, highly reproducible and having ability to detect higher level of polymorphism.
- A marker must to be polymorphic, i.e., it must exit in different forms so that chromosome carrying the mutant genes can be distinguished from the chromosomes with the normal gene by a marker it also carries.
- Genetic polymorphism is defined as the simultaneous occurrence of a trait in the same population of two discontinuous variants or genotypes.
- DNA markers segregate as single genes and they are not affected by the environment. DNA is easily extracted from plant
- materials and its analysis can be cost and labour effective. The first such DNA
- markers to be utilised were fragments produced by restriction digestion–the
- restriction fragment length polymorphism (RFLP) based genes marker.
An ideal molecular marker must have some desirable properties.
- 1. Highly polymorphic nature: It must be polymorphic as it is polymorphism that is measured for genetic diversity studies.
- 2. Codominant inheritance: Determination of homozygous and heterozygous states of diploid organisms.
- 3. Frequent occurrence in genome: A marker should be evenly and frequently distributed throughout the genome.
- 4. Selective neutral behaviours: The DNA sequences of any organism are neutral to environmental conditions or management practices.
- 5. Easy access (availability): It should be easy, fast and cheap to detect.
- 6. Easy and fast assay.
- 7. High reproducibility.
- 8. Easy exchange of data between laboratories.
Various types of molecular markers are utilized to evaluate DNA polymorphism and are generally classified as hybridization based markers and polymerase chain reaction (PCR)-based markers.
Classification of molecular markers
- Molecular markers are classified into various groups on the basis of
- (1) mode of gene action (co-dominant or dominant markers);
- (2) method of detection (hybridization-based molecular markers or polymerase chain reaction (PCR)- based markers);
- 3) mode of transmission (paternal organelle inheritance, maternal organelle inheritance, bi-parental nuclear inheritance or maternal nuclear inheritance)
| List of Markers | Acronym |
| Restriction Fragment Length Polymorphism | RFLP |
| Random Amplified Polymorphic DNA | RAPD |
| Amplified Fragment Length Polymorphism | AFLP |
| Variable Number Tandem Repeat | VNTR |
| Polymerase chain reaction (PCR)-sequencing | PCRS |
| Microsatellites or simple sequence repeat | SSR |
| Inter simple sequence repeats | (ISSR) |
| Single-strand conformation polymorphism | (SSCP) |
| Cleaved amplified polymorphic sequence | (CAPS) |
| Sequence characterised amplified region | (SCAR) |
| Single nucleotide polymorphism | (SNP) |
Restriction fragment length polymorphism (RFLP):
- RFLP was the very first technology employed for the detection of polymorphism, based on the DNA sequence differences. RFLP is mainly based on the altered restriction enzyme sites, as a result of mutations and re-combinations of genomic DNA
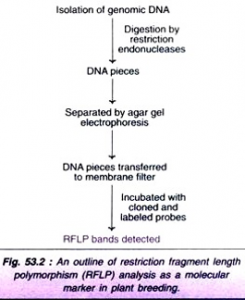
- The procedure basically involves the isolation of genomic DNA, its digestion by restriction enzymes, separation by electrophoresis, and finally hybridization by incubating with cloned and labeled probes

- Based on the presence of restriction sites, DNA fragments of different lengths can be generated by using different restriction enzymes.
- two DNA molecules from two plants (A and B) are shown. In plant A, a mutations has occurred leading to the loss of restriction site that can be digested by EcoRI.
- The result is that when the DNA molecules are digested by the enzyme Hindlll, there is no difference in the DNA fragments separated. However, with the enzyme EcoRI, plant A DNA molecules is not digested while plant B DNA molecule is digested. This results in a polymorphic pattern of separation.
Advantages
- RFLPs are generally found to be moderately polymorphic.
- In addition to their high genomic abundance and their random distribution, RFLPs have the advantages of showing codominant alleles and having high reproducibility.
- RFLPs, being codominant markers, can detect coupling phase of DNA molecules, as DNA fragments from all homologous chromosomes are detected.
- They are very reliable markers in linkage analysis and breeding and can easily determine if a linked trait is present in a homozygous or heterozygous state in individual, information highly desirable for recessive traits.
Disadvantages
- The of utility RFLPs has been hampered due to the large quantities (1–10 μg) of purified,
- high molecular weight DNA are required for each DNA digestion and Southern blotting.
- Larger quantities are needed for species with larger genomes and for the greater number of times needed to probe each blot.
- The requirement of radioactive isotope makes the analysis relatively expensive and hazardous.
- The assay is time-consuming and labour intensive and only one out of several markers may be polymorphic, which is highly inconvenient especially for crosses between closely related species.
- Their inability to detect single base changes restricts their use in detecting point mutations occurring within the regions at which they are detecting polymorphism.
Applications
- RFLPs can be applied in diversity and phylogenetic studies ranging from individuals within populations or species, to closely related species.
- RFLPs have been widely used in gene mapping studies because of their high genomic abundance due to the sample availability of different restriction enzymes and random distribution throughout the genome.
- They also have been used to investigate relationships of closely related taxa, as fingerprinting tools, for diversity studies and for studies of hybridization and introgression, including studies of gene flow between crops and weeds.
- RFLP markers were used for the first time in the construction of genetic maps by Botstein and others.
- A set of RFLP genetic markers provided the opportunity to develop a detailed genetic map of lettuce.
Random Amplified Polymorphic DNA (RAPD)
- RAPD is a molecular marker based on PCR amplification.
- The DNA isolated from the genome is denatured the template molecules are annealed with primers, and amplified by PCR.
- Single short oligonucleotide primers (usually a 10-base primer) can be arbitrarily selected and used for the amplification DNA segments of the genome (which may be in distributed throughout the genome).
- The amplified products are separated on electrophoresis and identified.

- Based on the nucleotide alterations in the genome, the polymorphisms of amplified DNA sequences differ which can be identified as bends on gel electrophoresis.
- Genomic DNA from two different plants often results in different amplification patterns i.e. RAPDs.
- This is based on the fact that a particular fragment of DNA may be generated from one individual, and not from others.
- This represents polymorphism and can be used as a molecular marker of a particular species.
Applications of RAPD Marker in Plants
- 1. RAPD is used to distinguish between variety is based on difference in DNA sequence.
- RAPD have been used to identify nearly 15 commercial sunflower varieties.
- A specific RAPD marker has been used to select for high and low β-glucan content between barley varieties.
- 2. RAPD markers are employed in the construction of genetic maps.
- Genetic maps of several plants including model plant Arabidopsis and tobacco have been constructed.
- RAPD markers have been used to construct 15 linkage groups in coffee.
- Both genomic and chloroplast DNA provided the source of probes.
- These markers also accelerate back crossing process and allow the selection of individually with more of recurrent genome at each generation facilitate breeding programme to be completed within few generations.
- 3. RAPD molecular marker used in the direct selection of desirable trait.
- Molecular marker linked to the trait of interest can be screened for at any stage in the breeding programme.
- 4. RAPD and other molecular markers have great value in the selection for desirable trait in long-lived species which takes long time for maturity and show phenotypic character.
- For example, avacado (Persea americana) fruit quality can be assessed in seedling itself using RAPD molecular marker.
- 5. RAPD markers have been used to identify several disease resistant genes in plants.
- The rp94 gene is responsible for resistance to stem rust (Puccinia gramnis) in barley.
- RAPD markers identified to link to this gene.
- Similarly, RAPD markers linked to heat smut resistance gene have been characterized.
- Controlling of height in barley plant by specific gene has been used to locate dwarfism gene by RAPD marker
- 6. In tissue culture work, somatic hybrids involving protoplast fusion requires thorough screening. However, screening of somatic hybrid is cumbersome. Therefore, RAPD markers can be exploited in identifying somatic hybrids. RAPD analysis provides an important tool for the characterization of biodiversity.
- Identification of areas rich in endemic genotypes helps in habitat conservative and prevents species extinction. Molecular analysis of genetic diversity using RAPD or RFLP in plant genetic germplasm collection facilitate better management especially space and resources are serious constraints.
- RAPD analysis has been used for the identification of duplicates in germplasm. These duplicates are then discarded once no morphological differences were detected. RAPD analysis has been implicated in the analysis of rice genome collections held at the International Rice Research Institute, Phillipines.
- Genetic diversity was carried out in a set a 63 tetraploid wheat genotype. Which comprises 24 duran land races, 18 duran cultivars and nine diococcum cultivars, two wild tetraploid species? The duran and dicoccum wheat genotypes are part of the germplasm used in Indian tetraploid wheat breeding programme.
- RAPD scoring analysis reveals 78% were polymorphic in different categories of Indian tetraploid wheat. These indicate that RAPD diversity data can be used in breeding improved cultivars and maintaining genetic diversity in germplasm. Similarly RAPD markers were generated from 6 groups of 23 varieties of Tibetian barley. Nearly 23 RAPD and 29 genes loci were identified on 72 chromosomes.
- RAPD have also been used in variety identification and purity in grain processing in food industry. For example, particular duran variety of wheat is used in the preparation of food products. Contamination of other variety can be identified by using these molecular markers.
Amplified Fragment Length Polymorphism (AFLP)
- AFLP is a novel technique involving a combination of RFLP and RAPD.
- AFLP is based on the principle of generation of DNA fragments using restriction enzymes and oligonucleotide adaptors (or linkers), and their amplification by PCR.
- Thus, this technique combines the usefulness of restriction digestion and PCR.
- The DNA of the genome is extracted.
- It is subjected to restriction digestion by two enzymes (a rare cutter e.g. Msel; a frequent cutter e.g. EcoRI).
- The cut ends on both sides are then ligated to known sequences of oligonucleotides

- PCR is now performed for the pre-selection of a fragment of DNA which has a single specific nucleotide.
- By this approach of pre-selective amplification, the pool of fragments can be reduced from the original mixture.
- In the second round of amplification by PCR, three nucleotide sequences are amplified.
- This further reduces the pool of DNA fragments to a manageable level (< 100).
- Autoradiography can be performed for the detection of DNA fragments.
- Use of radiolabeled primers and fluorescently labeled fragments quickens AFLP.
- AFLP is very sensitive and reproducible.
- It does not require prior knowledge of sequence information.
- By AFLP, a large number of polymorphic bands can be produced and detected.