RECOMBINATION
Introduction
- In the two preceding sections we discussed the mechanisms by which DNA sequences in cells are maintained from generation to generation with very little change.
- Although such genetic stability is crucial for the survival of individuals, in the longer term the survival of organisms may depend on genetic variation, through which they can adapt to a changing environment.
- Thus an important property of the DNA in cells is its ability to undergo rearrangements that can vary the particular combination of genes present in any individual genome, as well as the timing and the level of expression of these genes.
- These DNA rearrangements are caused by genetic recombination. Two broad classes of genetic recombination are commonly recognized : –
(a) General recombination and
(b) Site-specific recombination.
- In general recombination, genetic exchange takes place between any pair of homologous DNA sequences, usually located on two copies of the same chromosome.
- One of the most important examples is the exchange of sections of homologous chromosomes (homologues) in the course of meiosis.
- This “crossing-over” occurs between tightly apposed chromosomes early in the development of eggs and sperm and it allows different versions (alleles) of the same gene to be tested in new combinations with other genes, increasing the chance that at least some members of a mating population will survive in a changing environment.
- Although meiosis occurs only in eukaryotes, the advantage of this type of gene mixing is so great that mating and the reassortment of genes by general recombination are also widespread in bacteria.
- This process leads to offspring having different combinations of genes from their parents and can produce new chimeric alleles.
- Enzymes called recombinases catalyze natural recombination reactions. RecA, the recombinase found in E. coli, is responsible for the repair of DNA double strand breaks (DSBs).
- In yeast and other eukaryotic organisms there are two recombinases required for repairing DSBs. The RAD51 protein is required for mitotic and meiotic recombination and the DMC1 protein is specific to meiotic recombination.
- Chromosomal crossover refers to recombination between the paired chromosomes inherited from each of one’s parents, generally occurring during meiosis.
- During prophase-I the four available chromatids are in tight formation with one another. While in this formation, homologous sites on two chromatids can mesh with one another, and may exchange genetic information.
- Because recombination can occur with small probability at any location along chromosome, the frequency of recombination between two locations depends on their distance.
- Therefore, for genes sufficiently distant on the same chromosome the amount of crossover is high enough to destroy the correlation between alleles.
- In gene conversion, a section of genetic material is copied from one chromosome to another, but leaves the donating chromosome unchanged.
- Recombination can occur between DNA sequences that contain no sequence homology.
- This is referred to as Nonhomologous recombination or Nonhomologous end joining.
- DNA homology is not required in site-specific recombination.
- Instead, exchange occurs at short, specific nucleotide sequences (on either one or both of the two participating DNA molecules) that are recognized by a variety of site-specific recombination enzymes.
- Site-specific recombination therefore alters the relative positions of nucleotide sequences in genomes.
- In some cases these changes are scheduled and organized, as when an integrated bacterial virus is induced to leave a chromosome of a bacterium under stress; in others they are haphazard, as when the DNA sequence of a transposable element is inserted at a randomly selected site in a chromosome.
- As for DNA replication, most of what we know about the biochemistry of genetic recombination has come from studies of prokaryotic organisms, especially of E. coli and its viruses.
Independent Assortment and Crossing Over
Independent Assortment –
- The Law of Independent Assortment, also known as “Inheritance Law”, states that the inheritance pattern of one trait will not affect the inheritance pattern of another.
- While Mendel’s experiments with mixing one trait always resulted in a 3:1 ratio between dominant and recessive phenotypes, his experiments with mixing two traits (dihybrid cross) showed 9:3:3:1 ratios (Figure 3.1).
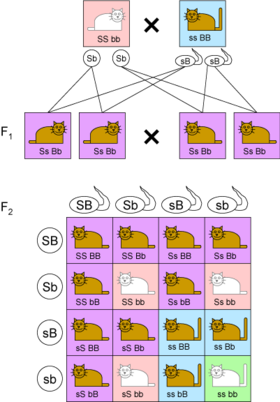
Figure 3.1: The genotypes of two independent traits show a 9:3:3:1 ratio in the F2 generation.
- In this example, coat color is indicated by B (brown, dominant) or b (white) while tail length is indicated by S (short, dominant) or s (long).
- When parents are homozygous for each trait (‘SSbb and ssBB), their children in the F1 generation are heterozygous at both loci and only show the dominant phenotypes.
- If the children mate with each other, in the F2 generation all combination of coat color and tail length occur: 9 are brown/short (purple boxes), 3 are white/short (pink boxes), 3 are brown/long (blue boxes) and 1 is white/long (green box).
- But the 9:3:3:1 table shows that each of the two genes is independently inherited with a 3:1 ratio.
- Mendel concluded that different traits are inherited independently of each other, so that there is no relation, for example, between a cat’s color and tail length.
- This is actually only true for genes that are not linked to each other. Independent assortment occurs during meiosis I in eukaryotic organisms, specifically anaphase I of meiosis, to produce a gamete with a mixture of the organism’s maternal and paternal chromosomes.
- Along with chromosomal crossover, this process aids in increasing genetic diversity by producing novel genetic combinations.
- Of the 46 chromosomes in a normal diploid human cell, half are maternally-derived (from the mother’s egg) and half are paternally-derived (from the father’s sperm).
- This occurs as sexual reproduction involves the fusion of two haploid gametes (the egg and sperm) to produce a new organism having the full complement of chromosomes.
- During gametogenesis – the production of new gametes by an adult – the normal complement of 46 chromosomes needs to be halved to 23 to ensure that the resulting haploid gamete can join with another gamete to produce a diploid organism.
- An error in the number of chromosomes, such as those caused by a diploid gamete joining with a haploid gamete, is termed aneuploidy.
- In independent assortment the chromosomes that end up in a newly-formed gamete are randomly sorted from all possible combinations of maternal and paternal chromosomes. Because gametes end up with a random mix instead of a pre-defined “set” from either parent, gametes are therefore considered assorted independently.
- As such, the gamete can end up with any combination of paternal or maternal chromosomes.
- Any of the possible combinations of gametes formed from maternal and paternal chromosomes will occur with equal frequency.
- For human gametes, with 23 pairs of chromosomes, the number of possibilities is 223 or 8,388,608 possible combinations.
- The gametes will normally end up with 23 chromosomes, but the origin of any particular one will be randomly selected from paternal or maternal chromosomes.
- This contributes to the genetic variability of progeny.
Crossing Over –
- Crossing over occurs between equivalent portions of two nonsister chromatids (Figure 3.2). Each chromatid contains a single molecule of DNA.
- So the problem of crossing over is really a problem of swapping portions of adjacent DNA molecules.
- It must be done with great precision so that neither chromatid gains or loses any genes. In fact, crossing over has to be sufficiently precise that not a single nucleotide is lost or added at the crossover point if it occurs within a gene.
- Otherwise a frameshift would result and the resulting gene would produce a defective product or, more likely, no product at all.
- How do nonsister chromatids ensure that crossing over between them will occur without the loss or gain of a single nucleotide?
- One plausible mechanism for which there is considerable laboratory evidence postulates the following events.
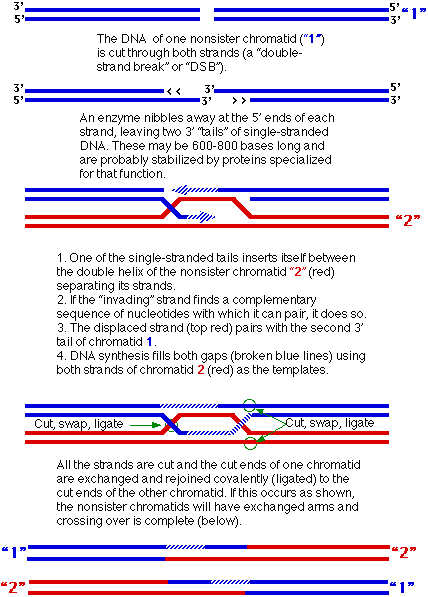
Figure 3.2: Mechanism of Crossing Over
- Note that each recombinant DNA molecule includes a region where nucleotides from one of the original molecules are paired with nucleotides from the other.
- But no matter, the need for a smooth double helix guarantees that each exchange takes places without any gain or loss of nucleotides.
- So long as the total number of nucleotides in each strand and the complementarities (A-T, C-G) are preserved, this “heteroduplex” region (which may extend for hundreds of base pairs) will only rarely have genetic consequences.
- And these may, in fact, be helpful because the synthesis of a short stretch of DNA using the template provided by the other chromatid also provides a mechanism for repairing any damage that might have been present on the “invading” strand of DNA.
- If the cut in the molecule 1 occurs in the region of a mutation, the damaged or incorrect nucleotides can be digested away. Refilling the resulting gap, using the undamaged molecule 2 as the template, repairs the damage to molecule 1.
- Why should the cutting and ligation be limited to the strands shown?
- They are not. Half the time the cutting and ligating rejoins the original parental arms.
- In these cases, no crossover takes place.
- The only genetic change that might have occurred is a transfer of some genetic information in the heteroduplex region.
- So crossing over not only provides a mechanism for genetic recombination during meiosis but also provides a means of repairing damage to the genome.



