- Homologous chromosomes start pairing at one or more points along the lengths of chromosomes during this stage.
- Pairing extends along the rest of the chromosome length in a zipper like fashion.
- Although heredity, temperature, nutrition, etc. are known to affect pairing, the mechanism of pairing is not well understood.
- Pairing or synapsis is a very precise process in which only homologous parts of chromosomes come together.
- Non-homologous parts are looped out and thus remain unpaired.
- A pair of homologous chromosomes is called bivalent .
- At any one point only two chromosomes can get paired.
- This is evident when three or four homologous chromosomes are held together to form multivalent, but even then at any given point only two chromosomes are paired.
- Under the electron microscope the synaptonemal complex appears multilayered.
- There is a central axis which is flanked on both the sides by an axial element.
- There is a central space in between the central and axial elements.
- Chromatin is dispersed towards the outer faces of the axial element.
- DNase destroys the chromatin but not the axial or central elements.
- These are destroyed by trypsin, indicating thereby that both the central and axial elements are proteinaceous in nature.
- The central axis is composed of a substance which is synthesized during pairing.
- Synaptonemal complexes not formed if the homologous are experimentally prevented from pairing.
- The ends of chromosomes are attached to the inner surface of the nuclear membrane at this stage.
- A part of the DNA (about 0.3%in lilium) that remains unreplicated during interphase is replicated during this stage of meiosis
Stages of Chromosomal Pairing
Chromosomal pairing proceeds through several stages:
1. Recognition and Alignment of Homologs
- Homologous chromosomes recognize each other through DNA sequence homology.
- This process is guided by DNA double-strand breaks (DSBs) introduced intentionally by Spo11 enzyme.
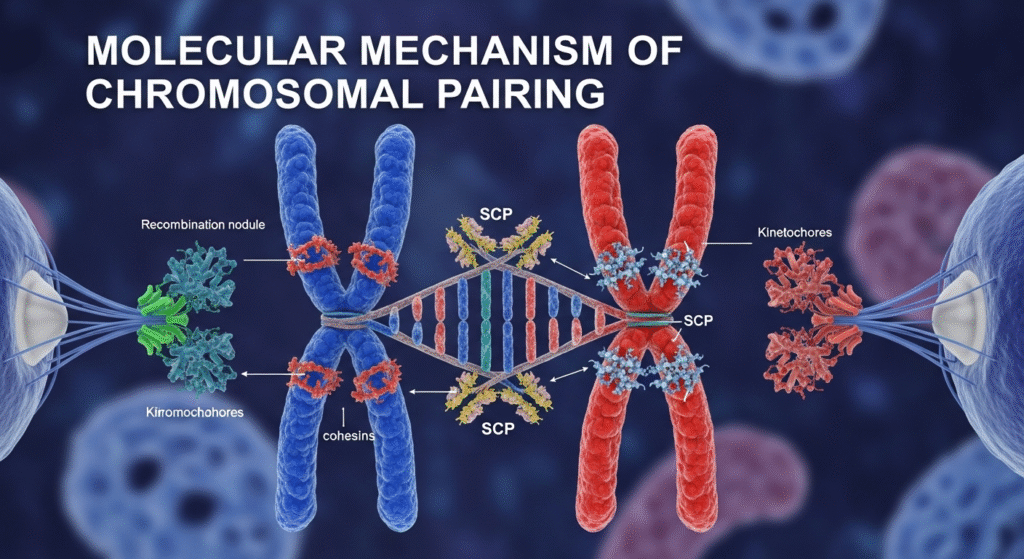
- The cohesin complex helps hold sister chromatids together, stabilizing the chromosomes during alignment.
2. Homology Search
- Recombinase proteins, such as Rad51 and Dmc1, are loaded onto the single-stranded DNA overhangs at the DSB sites.
- These proteins mediate a homology search by aligning the broken DNA with homologous sequences on the non-sister chromatid.
3. Synapsis (Formation of Synaptonemal Complex)
- Once homologs align, a protein structure called the Synaptonemal Complex (SC) forms between them.
- The SC consists of:
- Lateral elements (along each homolog)
- Central element
- Transverse filaments (connect lateral to central elements)
- Lateral elements (along each homolog)
- This complex holds homologs in tight association, ensuring precise recombination.
4. Recombination (Crossing Over)
- Homologous recombination occurs at points of contact called chiasmata.
- Proteins like MLH1, MSH4, and MSH5 are involved in crossover formation.
- Crossovers ensure genetic variability and correct tension on homologs for metaphase alignment.
Key Molecular Players in Chromosomal Pairing
| Molecule/Structure | Function |
| Spo11 | Creates DSBs for initiating recombination |
| Rad51, Dmc1 | Catalyze strand invasion and homology search |
| Cohesin | Maintains sister chromatid cohesion |
| Synaptonemal Complex (SC) | Brings homologs into close alignment |
| ZYP1 (in plants), SYCP1/SYCP2/SYCP3 (in animals) | Structural proteins of the SC |
| Mismatch repair proteins (MSH4/MSH5) | Stabilize homologous interactions |
| MLH1, MLH3 | Facilitate crossover formation |
| ATM, ATR kinases | DNA damage response, monitor DSB repair |
Outcomes of Proper Chromosomal Pairing
- Accurate chromosome segregation
- Prevention of aneuploidy
- Increased genetic diversity via recombination
- Proper gamete formation
Errors in Pairing: Consequences
Improper or failed pairing may lead to:
- Non-disjunction (leading to trisomy or monosomy)
- Infertility
- Genetic disorders (e.g., Down syndrome)
- Meiotic arrest or apoptosis of gametes
MCQs: Molecular Mechanism of Chromosomal Pairing
🔹 Basic Concepts
- Which stage of meiosis involves chromosomal pairing?
a) Metaphase I
b) Anaphase I
c) Prophase I ✅
d) Telophase I - The term “bivalent” refers to:
a) Two non-homologous chromosomes
b) A pair of homologous chromosomes ✅
c) A duplicated chromosome
d) Sister chromatids - The pairing of homologous chromosomes occurs in which sub-stage of prophase I?
a) Leptotene
b) Zygotene ✅
c) Pachytene
d) Diplotene - Which enzyme initiates double-strand breaks (DSBs) during pairing?
a) Topoisomerase
b) Helicase
c) Ligase
d) Spo11 ✅ - Homologous pairing is initiated by:
a) Crossing over
b) Random alignment
c) DNA sequence homology ✅
d) Chromosome condensation
🔹 Molecular Mechanism
- The protein that helps maintain cohesion between sister chromatids is:
a) Tubulin
b) Kinase
c) Cohesin ✅
d) Myosin - The search for homologous DNA sequences is performed by:
a) Histones
b) Rad51 and Dmc1 ✅
c) DNA ligase
d) Polymerase - What structure forms to tightly associate homologous chromosomes?
a) Chiasmata
b) Centriole
c) Synaptonemal Complex (SC) ✅
d) Kinetochores - The synaptonemal complex is visible under:
a) Light microscope
b) Dissecting microscope
c) Electron microscope ✅
d) Phase contrast microscope - Which element of SC connects lateral elements?
a) Centrosome
b) Chromatin
c) Transverse filaments ✅
d) Histone tails
🔹 Proteins & Structures
- SYCP1, SYCP2, and SYCP3 are components of SC in:
a) Plants
b) Animals ✅
c) Fungi
d) Bacteria - ZYP1 is a component of synaptonemal complex in:
a) Plants ✅
b) Animals
c) Humans
d) Yeast - The central axis of SC is composed of:
a) DNA
b) RNA
c) Protein ✅
d) Lipids - DNase destroys:
a) SC proteins
b) Central elements
c) Chromatin ✅
d) Axial elements - Trypsin destroys:
a) DNA only
b) Chromatin
c) Axial and central SC elements ✅
d) Membrane
🔹 Recombination & Crossing Over
- Crossing over occurs at:
a) Spindle poles
b) Chiasmata ✅
c) Centromere
d) Nucleolus - Which proteins facilitate crossover?
a) SYCP2 & SYCP3
b) Rad51 & Spo11
c) MLH1 & MLH3 ✅
d) DNase & Dmc1 - Mismatch repair proteins involved in pairing are:
a) MLH1, RAD51
b) MSH4, MSH5 ✅
c) SYCP1, ZYP1
d) ATR, ATM - What ensures tension during metaphase alignment?
a) DNA replication
b) Crossovers ✅
c) Sister chromatid cohesion
d) Protein synthesis - Which kinase proteins monitor DSB repair?
a) CDKs
b) Cyclins
c) ATM, ATR ✅
d) Histones
🔹 Advanced Concepts
- Which chromosome ends attach to the nuclear membrane during pairing?
a) Middle arms
b) Centromeres
c) Telomeres ✅
d) Chiasmata - The zipper-like extension of pairing is observed in:
a) Cytokinesis
b) Kinetochore movement
c) Synapsis ✅
d) Telophase - What remains unpaired during homologous pairing?
a) Sister chromatids
b) Homologous DNA
c) Non-homologous chromosome regions ✅
d) Entire chromosome - The portion of DNA that replicates during pairing is approximately:
a) 2%
b) 1.5%
c) 5%
d) 0.3% (in Lilium) ✅ - What is the key function of the synaptonemal complex?
a) Break DNA
b) Condense chromatin
c) Facilitate alignment and recombination ✅
d) Activate centrioles
🔹 PYQs & Application (Based on UPSC/CSIR/NEET patterns)
- (NEET 2019) The point of crossing over between homologous chromosomes is:
a) Centromere
b) Telomere
c) Chiasma ✅
d) Locus - (CSIR NET) Which of the following recombinase proteins is meiosis-specific?
a) RAD51
b) Ligase
c) Polymerase
d) DMC1 ✅ - (NEET 2021) Cohesin proteins are important for:
a) Chromosome movement
b) Sister chromatid cohesion ✅
c) Spindle attachment
d) SC formation - (UPSC) Which protein is essential for initiating double-strand break in meiosis?
a) DMC1
b) MLH3
c) Spo11 ✅
d) ATM - (CUET 2023) The structure NOT part of synaptonemal complex is:
a) Lateral element
b) Transverse filament
c) Central element
d) Centromere ✅
🔹 Errors in Pairing
- Non-disjunction results from:
a) Proper crossing over
b) DNA replication
c) Improper pairing ✅
d) Telophase delay - Which genetic disorder is due to chromosome missegregation?
a) Hemophilia
b) Down syndrome ✅
c) Thalassemia
d) Color blindness - Infertility in males can result from errors in:
a) Translation
b) Replication
c) Chromosomal pairing ✅
d) Spindle assembly - Multivalent chromosome structures arise due to:
a) Polytene chromosomes
b) Crossing over
c) Multiple homologs pairing partially ✅
d) Lack of centromeres - If homologous chromosomes don’t pair, SC is:
a) Elongated
b) Not formed ✅
c) Modified
d) Divided
🔹 Research Insights / Facts
- The SC appears as how many layers under EM?
a) Single
b) Multilayered ✅
c) Bilayer
d) Diffuse - Which macromolecule forms axial and central elements?
a) DNA
b) RNA
c) Protein ✅
d) Lipid - Which repair protein complex detects mismatches in pairing?
a) Rad51
b) MSH4/MSH5 ✅
c) SYCP3
d) Cohesin - Which of the following is NOT part of SC?
a) Transverse filament
b) Central space
c) Kinetochore ✅
d) Lateral element - Protein synthesis specific to SC happens during:
a) Anaphase
b) Zygotene ✅
c) Leptotene
d) S-phase
🔹 True Statements
- Only two chromosomes can pair at one point even in multivalent structures. ✅
- Chromatin faces outwards from the SC axial element. ✅
- Pairing is influenced by heredity, nutrition, and temperature. ✅
- SC formation requires prior chromosome recognition. ✅
- Spo11 function is conserved in many organisms. ✅
🔹 Miscellaneous
- What is looped out during homologous pairing?
a) Homologous DNA
b) Non-homologous DNA ✅
c) Spindle fiber
d) Synaptonemal core - The final outcome of accurate pairing is:
a) Cancer
b) Mutation
c) Fertile gametes ✅
d) Apoptosis - Which structure stabilizes the alignment of homologs?
a) Centrosome
b) Synaptonemal Complex ✅
c) Golgi body
d) Actin - Which protein is NOT involved in recombination?
a) MSH5
b) MLH1
c) RAD51
d) Tubulin ✅ - What happens to gametes when chromosomal pairing fails?
a) Rapid cell division
b) Overgrowth
c) Apoptosis or arrest ✅
d) Multiplication